GENETICS: THE ROLE OF TRANSFER RNA AND THE CONTROL OF GENE EXPRESSION.
Transfer RNA molecules are smaller than mRNA molecules,
containing only about 75 to 80 nucleotides. Each consists of a single strand of
nucleic acid folded back on itself to form a ‘clover leaf’ shape. Transfer RNA
molecules bring specific amino acids from the cytoplasm to the ribosome so that
they attach to the growing polpeptide.
The Process by which an amino acid binds to a tRNA molecule is controlled by an enzyme. The process also involves the splitting of ATP. This is important because ATP gives the tRNA-amino acid complex enough energy to form a peptide bond when the amino acid is added to the growing polypeptide.
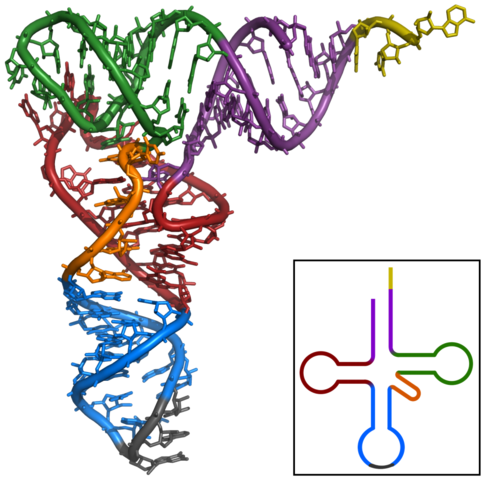
At one end of the tRNA molecule is the anticodon, a three-letter base sequence that matches the codon on the mRNA molecule. At the other end is a particular amino acid. So, for instance, if the codon on the mRNA reads AUG, which codes for the amino acid methionine, it needs a tRNA moleculce with an anticodon of UAC which arrives at the ribosome carrying a methionine molecule.
There are 64 different codons, but three of them code for ‘stop translating’. When such a codon arrives at the ribosome, no tRNA is needed. This means that tRNA molecules must match with 61 different codons. There are only 20 different amino acids, so some amino acids are translated from more than one codon. For example, the codons GGG, GGA, GGC and GGU all code for the amino acid glycine.
Like messenger RNA, transfer RNA is also made by transcription. In eukaryote DNA there are many genes whose sole function is to make tRNA. The DNA code is said to be degenerate, which means that there can be more than one codon for each individual amino acid. In any cell, most genes occur only once, but there are hundreds of genes that code for tRNA synthesis, to provide the vast numbers of these workhorse molecules that are needed. That tRNA genes do not code for proteins is another exception to the central dogma.
Polysomes and the rate of translation
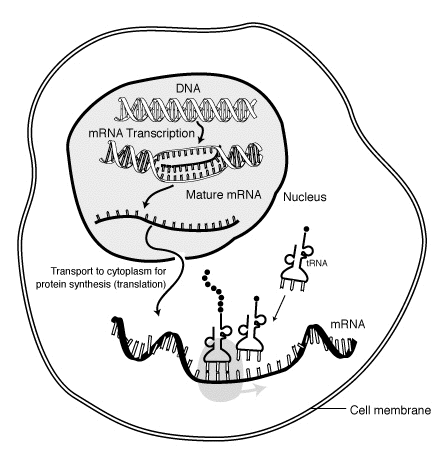
We have seen that the mRNA molecule is a long single strand of nucleotides and that the code is translated into a polypeptide at the point of contact with a ribosome. To achieve protein synthesis at a reasonable speed, ribosomes occur in clusters, called polysomes or polyribosomes, which all translate a different bit of the mRNA at the same time.
A JOURNEY ALONG A CHROMOSOME
If one single chromosome were unravelled, like pulling the wool out of a piece of kniting, we would be left with one long DNA strand. Although, this is one single molecule, it is several centimetres long and contains up to 4 000 genes.
How do you go about investigating such a molecule? One way is to work backwards: if you can isolate the mRNA molecules that are in the cytoplasm, you can trace their origin in the DNA. A clever technique called nucleic acid hybridisation is used to pinpoint the origin of the mRNA, giving the location of the gene itself. In this technique, DNA strands are denatured: when heated to 87 °C. Hydrogen honds that hold the two strands together are broken, and the individual strands separate. If messenger RNA strands from the cytoplasm are added to the mixture, they stick to the region of DNA on which they were originally made.
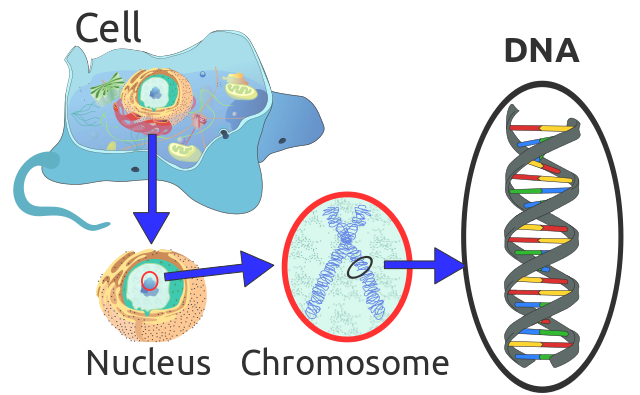
Location of eukaryote nuclear DNA within the chromosomes. Radio89, CC BY-SA 3.0
Studies using this technique have unexpectedly revealed that there is some non-coding DNA in genes. Most eukaryote genes contain regions, called introns, which do not find their way into the mRNA molecule at all. The parts of the gene that are expressed in the mRNA are called exons because they are the expressed regions of the gene.
Generally, a gene consists of a DNA sequence which is present only once.The non-coding sequences, however, tend to consist of repeated or ‘stuttered’ sequences. Humans all have the same repeated sequence, but in different individuals it is repeated a different number of times and in different places. This is the basis of DNA profiling.
The function of non-coding DNA is not known. It is probably not useless: much of the repeated DNA in a chromosome occurs around the region of its centromere and may have an important role in maintaining the physical stability of the chromosome as a whole. The DNA code can be compared to a page of writing in a book. If the letters were strung together continuously, we would find it difficult to read: the gaps between words help us to make sense of the sentences.
THE CONTROL OF GENE EXPRESSION
Most multicellular organisms start life as a zygote: a single fertilised cell. This cell has a complete set of genes. As the cell divides by mitosis, the DNA is copied faithfully, so each new cell also contains the same complete set of genes. How, then, do cells differentiate? How, for example, do some animal cells specialize into muscle, nerve or skin?
The answer lies in gene expression – how different genes are ‘switched on’, or expressed, in different cells.

At any one time, the average cell is probably using only 1 per cent of its available genes. Some of the expressed genes are used for ‘housekeeping’ – they code for the proteins needed by all cells, such as the enzymes involved in respiration. In contrast, other genes are expressed only in specialized cells. The genes for making insulin, for example, are expressed only in the β cells of the islets of Langerhans in the pancreas.
CONTROLLING TRANSCRIPTION
The control of transcription is the key to the selective activation of genes. A gene is switched on, or induced, when transcription begins. Transcription starts when the enzyme RNA polymerase and several proteins known as transcription factors bind to an area ‘upstream’ of the gene called the promoter region. The proteins assemble into a transcription initiation complex (TIC). Once this complex is in place, transcription can begin.
Some transcription factors are always present in a cell, some are present but in an inactive form, and some are not made until a key stage in a cell’s life. Genes remain switched off until the correct transcription factors are present in their active form. Transcription factors can be activated by signal proteins which may be hormones, growth factors or other regulatory molecules, or by environmental signals.
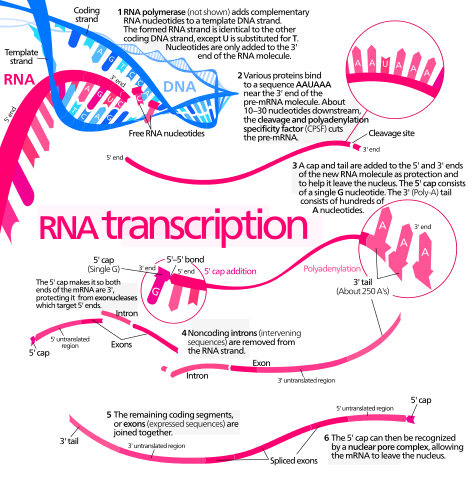
Simplified diagram of mRNA synthesis and processing. Enzymes not shown. Kelvinsong, CC BY 3.0
In addition, transcription can be prevented by the presence of certain repressor molecules, which attach to the promoter region and prevent the formation of the TIC.
Pioneering work on gene induction was done by Jacob and Monod in the early 1960s. They worked on the bacterium Escherichia coli. E. coli can digest lactose (milk sugar) by making the enzyme β galactosidase. When there is no lactose, the enzyme is not made because the β galactosidase gene is not active. However, the presence of lactose causes the gene to be induced. Jacob and Monod found that in the absence of lactose there was a repressor molecule that bound to the promotor region and prevented the formation of the TIC. When lactose was present, the repressor molecule was removed and the gene was activated. If we could control gene expression, we would be able to make new tissues and organs for repair and transplant.
Gene induction in humans happens all the time and it is extraordinarily complex. One set of genes that have been studied in detail are the stress response genes. There are lots of these in the genome and normally they are inactive. If a cell becomes stressed – because it is exposed to a carcinogen, or a high salt concentration, or because it gets short of oxygen, these genes are then induced to try to rescue the cell. Many of them code for enzymes that repair cell structures. Researchers in Cambridge in the UK have shown that when human cells in culture are stressed they start to express about 20 of these stress genes very quickly.
During development of the embryo, different genes are induced at different times and working out what genes are active and how they are activated is one of the great challenges left in biology. Cells in a developing embryo ‘know’ where they are because of chemical signals they receive from surrounding cells. These signals also control whether each cell divides or differentiates.
siRNA
This is a class of RNA molecules that has a variety of functions in the cell, including the control of transcription. They can act as repressor molecules because they prevent gene expression. Called small interfering RNA (siRNA) they are a class of small molecules, 20-25 nucleotides long and double-stranded. Most notably, siRNA is involved in the RNA interference (RNAi) pathway, where the siRNA interferes with the expression of a specific gene.
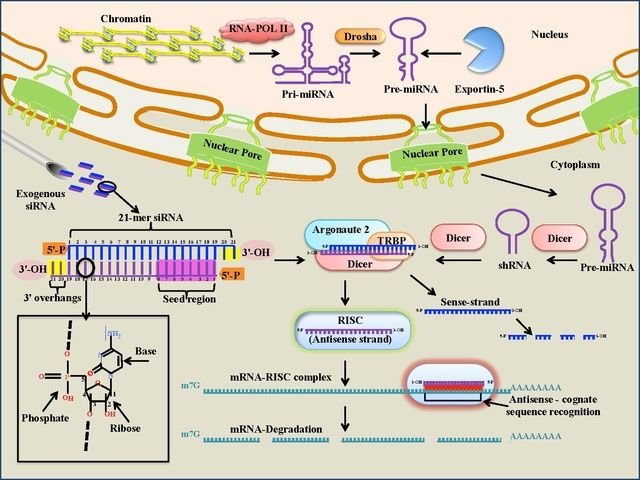
siRNAs have great potential in medicine and genetic engineering, because they can be made artificially and introduced into cells to bring about the specific knockdown of a particular gene, e.g. one that causes a particular type of cancer. Essentially, any gene of which the sequence is known can be de-activated by a tailor-made siRNA molecule.
DNA REPLICATION
Whenever a cell divides, the DNA must be copied. Otherwise, there would be no reproduction and no growth. When cells divide, each new daughter cell must have a complete set of genes. Replication, the mechanism of DNA copying, is therefore of fundamental importance.
Just three years after Crick and Watson published their model of DNA structure in the journal Nature, their theory about DNA replication was confirmed by Arthur Kornberg. He put DNA into a mixture containing all four nucleotides (each of the four bases attached to a phosphate and a sugar), together with the enzyme DNA polymerase, and showed that the DNA could replicate without any other factors present.
In the mixture, each DNA strand unwinds and acts as a template for the connstruction of a new strand. The exposed strand acts as a template on which the free nucleotides arrange themselves in exactly the same sequence as the intact strand they replace. This model is called semi-conservative replication because each of the resulting strands of DNA contains one strand from the original DNA and one newly synthesized strand: half has been conserved and half is new.
Strong support for the semi-conservative model of DNA replication came from the work of Meselson and Stahl. By growing bacteria with bases containing the radioisotope 15N (sometimes called heavy nitrogen) and then following its progress, they showed that each new DNA strand contains the original strand.
THE MECHANISM OF SEMI-CONSERVATIVE REPLICATION
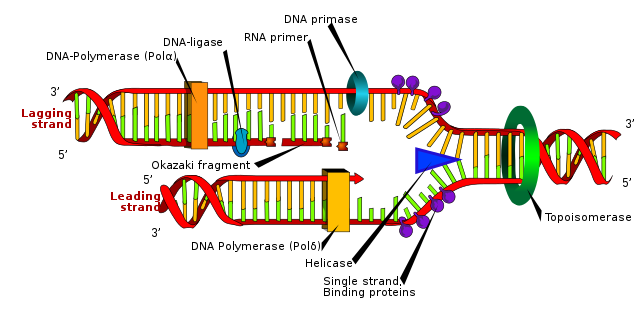
The basic mechanism of DNA replication is shown below. DNA is copied before the cell divides and the basic idea is simple: the helix unwinds and each strand acts as a template for the manufacture of a new, identical strand.
In practice, DNA replication is rather more complex than it appears, and is a good illustration of the importance of enzymes. Two helicases use energy from ATP to separate the DNA strands. Next, DNA binding proteins attach to keep the strands separate. DNA polymerase enzymes then move along the exposed strands, synthesising the new strands, often in short segments. Finally, DNA ligase enzymes join the segments together, completing the new DNA strands.
DNA proofreading
Mistakes in DNA replication occur regularly. The chances of the wrong base being added are between 1 in 108 and 1 in 1012. This may seem low, but so many nucleotides are added that this rate means approximately one mistake occurs for every ten genes that are copied. Clearly, such a rate would cause the death of most organisms. This disaster is prevented by the polymerase enzymes themselves. They can detect when the wrong base has been inserted, and pause to correct the mistake. In this way 99.9 per cent of mistakes are corrected. Uncorrected mistakes give rise to mutations.
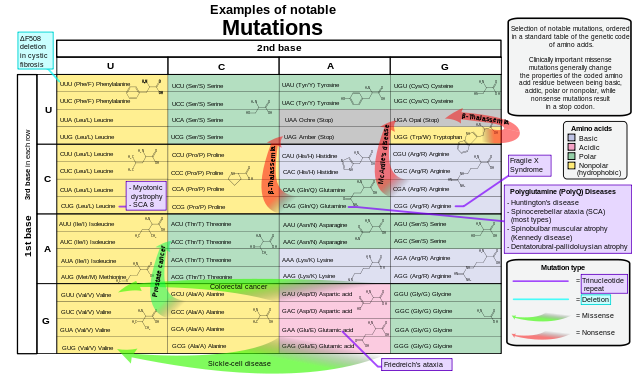
Examples of notable mutations that can occur in humans. Mikael Häggström, Public Domain
There are many different DNA repair mechanisms in cells to put right the genetic damage that inevitably accumulates during the life of an organism. The condition xeroderma pigmentosum illustrates the importance of these mechanisms: those affected are liable to get skin cancer because their cells cannot repair genetic damage caused by exposure to ultraviolet light.
Thanks for reading.
REFERENCES
https://en.wikipedia.org/wiki/Gene_expression
https://biologydictionary.net/trna/
https://www.ncbi.nlm.nih.gov/books/NBK21603/
https://en.wikipedia.org/wiki/Transfer_RNA
https://www.nature.com/scitable/definition/trna-transfer-rna-256
https://en.wikipedia.org/wiki/Polysome
https://www.sciencedirect.com/topics/medicine-and-dentistry/polysome
https://www.ncbi.nlm.nih.gov/pubmed/26965262
https://academic.oup.com/nar/article/45/3/e15/2972201
https://www.pnas.org/content/115/17/4411
https://stevemorse.org/genetealogy/beyond.htm
https://www.rigb.org/christmas-lectures/2013-life-fantastic/chromosome-videos
https://en.wikipedia.org/wiki/Nucleic_acid_hybridization
https://www.medicinenet.com/script/main/art.asp?articlekey=14349
https://themedicalbiochemistrypage.org/gene-regulation.php
https://en.wikipedia.org/wiki/Regulation_of_gene_expression
http://sphweb.bumc.bu.edu/otlt/MPH-Modules/PH/DNA-Genetics/DNA-Genetics7.html
https://www.ncbi.nlm.nih.gov/pubmed/20569216
This post has been voted on by the SteemSTEM curation team and voting trail. It is elligible for support from @curie and @minnowbooster.
If you appreciate the work we are doing, then consider supporting our witness @stem.witness. Additional witness support to the curie witness would be appreciated as well.
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
Thanks for having used the steemstem.io app and included @steemstem in the list of beneficiaries of this post. This granted you a stronger support from SteemSTEM.
Hello,
Your post has been manually curated by a @stem.steem curator.
We are dedicated to supporting great content, like yours on the STEMGeeks tribe.
Please join us on discord.