🔥#MESScience 4: Review of COVID-19 "Virus" Isolation Paper
In #MESScience 4 I go over a thorough review of the main paper claiming isolation of the "virus" that causes COVID-19. I also go over an overview of the mainstream narrative on COVID-19 and the supposed cases and death figures. I ask the artificial intelligence (AI) chatbot ChatGPT to give me a link to the "main paper" on COVID-19 "virus" isolation, to which it gives out the paper titled "A Novel Coronavirus from Patients with Pneumonia in China, 2019" by Zhu et al published on January 24, 2020 in The New England Journal of Medicine: https://www.nejm.org/doi/full/10.1056/NEJMoa2001017
I go over every sentence of the above paper, provide context, and expand on technical terms where necessary. I also go over an overview of the mainstream explanation on nucleic acids, genomes, and cell theory to better grasp the claims in the paper. In the paper, a handful of patients from Wuhan, China on December 2019 reported pneumonia (or inflammation of the lungs) of "unknown cause", to which the China Center of Disease Control (China CDC) sent a rapid response team to investigate. The paper outlines the results of the investigation.
After examining the paper in detail, it is clear that there are more assumptions than actual common sense isolation of a "virus". The paper specifies genetic sequencing technology was used as the basis for their claim of a new "virus", and they do this by looking for common sequences of other "coronaviruses" in global databases. The physical laboratory experiments performed involved taking patient airway lung fluid, then mixing with chemicals, centrifuging, and assuming the supernatant has the "virus" inside. The supernatant and cell cultures were then heavily processed, stained with heavy metals, embedded in resin, cut in ultrathin sections, and then placed in high-powered electron microscopes. The results show blurry, dead, black and white particles, which are assumed to be the "virus". In my opinion, the paper was not convincing in the slightest that any actual particle was "isolated", and much less a "disease causing virus". Further study will be required to examine the genetic sequencing science that is used to claim the existence of these so-called "viruses".
The topics covered as well as their timestamps are listed below.
- Introduction: 0:00
- #MESScience Video Series: 1:25
- MES Research Note: 2:51
- Topics to Cover: 3:25
- Mainstream Overview of COVID-19: 5:10
- ChatGPT on the Main Paper on COVID Virus Isolation: 18:47
- Paper: A Novel Coronavirus from Patients with Pneumonia in China, 2019: 21:37
- Paper Abstract: 23:30
- Summary: 33:32
- Paper Introduction: 34:40
- Summary: 46:31
- Quick Overview of Nucleic Acids and Genomes: 48:26
- Paper Methods
- Detecting "Viruses": 58:43
- Summary: 1:10:07
- Isolating the "Virus": 1:13:04
- Summary: 1:33:39
- Electron Microscope Preparation: 1:37:01
- Summary: 1:46:11
- Sequencing the "Virus": 1:48:11
- Summary: 1:55:43
- RNA Assumed to be from a "Virus": 1:56:48
- Summary: 2:00:09
- Detecting "Viruses": 58:43
- Paper Results
- Patient Results: 2:01:34
- Summary: 2:09:16
- Detection and Isolation of a New "Virus": 2:10:57
- Summary: 2:27:33
- Genetic Characterization of the "Virus": 2:33:19
- Summary: 2:42:35
- Patient Results: 2:01:34
- Paper Discussion: 2:44:08
- Summary: 2:55:01
- MES Summary: 2:58:10
- MES Conclusions: 3:10:08
Stay tune for #MESScience Part 5...
Download Video Notes: https://1drv.ms/b/s!As32ynv0LoaIieFrmN8-j483bZv0OQ?e=MiN3gi
View video notes on the Hive blockchain: https://peakd.com/hive-128780/@mes/messcience-4-review-of-covid-19-virus-isolation-paper
View video sections playlist: https://www.youtube.com/playlist?list=PLai3U8-WIK0E4aQ_cq4ZDD2WGiDU5vVgx
View the full #MESScience series: http://mes.fm/science-playlist
Watch video on:
- 3Speak: https://3speak.tv/watch?v=mes/nffierng
- Odysee: https://odysee.com/@mes:8/MESScience-4-Review-of-COVID-19-Virus-Isolation-Paper:a
- BitChute: https://www.bitchute.com/video/UmzzyzSXT0Oi/
- Rumble: https://rumble.com/v2s9cdm-messcience-4-review-of-covid-19-virus-isolation-paper.html
- DTube: https://d.tube/#!/v/mes/8h9wphs754v
- YouTube: https://youtu.be/cvDO85Rw4d8
View Video Notes Below!
Download these notes: Link is in video description.
View these notes as an article: https://peakd.com/@mes
Subscribe via email: http://mes.fm/subscribe
Donate! :) https://mes.fm/donate
Buy MES merchandise! https://mes.fm/store
More links: https://linktr.ee/matheasy
Follow my research in real-time on my MES Links Telegram: https://t.me/meslinks
Subscribe to MES Truth: https://mes.fm/truthReuse of my videos:
- Feel free to make use of / re-upload / monetize my videos as long as you provide a link to the original video.
Fight back against censorship:
- Bookmark sites/channels/accounts and check periodically
- Remember to always archive website pages in case they get deleted/changed.
Recommended Books:
- "Where Did the Towers Go?" by Dr. Judy Wood: https://mes.fm/judywoodbook
Join my forums!
- Hive community: https://peakd.com/c/hive-128780
- Reddit: https://reddit.com/r/AMAZINGMathStuff
- Discord: https://mes.fm/chatroom
Follow along my epic video series:
- #MESScience: https://mes.fm/science-playlist
- #MESExperiments: https://peakd.com/mesexperiments/@mes/list
- #AntiGravity: https://peakd.com/antigravity/@mes/series
-- See Part 6 for my Self Appointed PhD and #MESDuality breakthrough concept!- #FreeEnergy: https://mes.fm/freeenergy-playlist
- #PG (YouTube-deleted series): https://peakd.com/pg/@mes/videos
NOTE #1: If you don't have time to watch this whole video:
- Skip to the end for Summary and Conclusions (if available)
- Play this video at a faster speed.
-- TOP SECRET LIFE HACK: Your brain gets used to faster speed!
-- MES tutorial: https://peakd.com/video/@mes/play-videos-at-faster-or-slower-speeds-on-any-website- Download and read video notes.
- Read notes on the Hive blockchain #Hive
- Watch the video in parts.
-- Timestamps of all parts are in the description.Browser extension recommendations:
- Increase video speed: https://mes.fm/videospeed-extension
- Increase video audio: https://mes.fm/volume-extension
- Text to speech: https://mes.fm/speech-extension
--Android app: https://mes.fm/speech-android
🔥#MESScience 4: Review of COVID-19 "Virus" Isolation Paper

#MESScience Video Series
Here is a list of the MES Science videos that I have done so far, which include links to the corresponding Hive posts and YouTube sections playlists:
MES Science Tutorials
- YouTube playlist short URL: mes.fm/science-playlist
- How Does a Powerball Gyroscope Work? + Gyros are Inverted Pendulums
- Vortex Math Part 1: Number Theory and Modular Arithmetic
- Overview of Biology
- Review of COVID-19 Virus Isolation Paper (Current Video)
MES Research Note
In this video, I go over a methodical examination of the main paper on COVID "virus" isolation.
The reason for this methodical approach is that it helps me to learn and understand the paper while also presenting it to the public.
So, while it may be a long and tedious video, I believe it will better crystalize the mainstream science involved in the fear-mongering of a new "deadly virus" that requires us to obey "health authorities".
And with that, buckle up folks!
Topics to Cover
- Mainstream Overview of COVID-19
- ChatGPT on the Main Paper on COVID Virus Isolation
- Paper: A Novel Coronavirus from Patients with Pneumonia in China, 2019
- Paper Abstract
- Summary
- Paper Introduction
- Summary
- Quick Overview of Nucleic Acids and Genomes
- Paper Methods:
- Detecting "Viruses"
- Summary
- Isolating the "Virus"
- Summary
- Electron Microscope Preparation
- Summary
- Sequencing the "Virus"
- Summary
- RNA Assumed to be from a "Virus"
- Summary
- Detecting "Viruses"
- Paper Results
- Patient Results
- Summary
- Detection and Isolation of a New "Virus"
- Summary
- Genetic Characterization of the "Virus"
- Summary
- Patient Results
- Paper Discussion
- Summary
- MES Summary
- MES Conclusions
Mainstream Overview of COVID-19
Let's first go over the mainstream overview of COVID-19, as summarized by the main mainstream narrative machine: Wikipedia.
https://en.wikipedia.org/wiki/COVID-19
Retrieved: 12 March 2023
Archive: https://archive.ph/wip/2IxfpCoronavirus disease 2019 (COVID-19) is a contagious disease caused by a virus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first known case was identified in Wuhan, China, in December 2019.[5] The disease quickly spread worldwide, resulting in the COVID-19 pandemic.
The symptoms of COVID‑19 are variable but often include fever,[6] cough, headache,[7] fatigue, breathing difficulties, loss of smell, and loss of taste.[8][9][10] Symptoms may begin one to fourteen days after exposure to the virus. At least a third of people who are infected do not develop noticeable symptoms.[11] Of those who develop symptoms noticeable enough to be classified as patients, most (81%) develop mild to moderate symptoms (up to mild pneumonia), while 14% develop severe symptoms (dyspnea, hypoxia, or more than 50% lung involvement on imaging), and 5% develop critical symptoms (respiratory failure, shock, or multiorgan dysfunction).[12] Older people are at a higher risk of developing severe symptoms. Some people continue to experience a range of effects (long COVID) for months after recovery, and damage to organs has been observed.[13] Multi-year studies are underway to further investigate the long-term effects of the disease.[13]
Frequency: 676,609,955 cases
Deaths: 6,881,955
Note that the world population is currently at about 8.021 billion.
Here is a % breakdown I made of the above figures.
Microsoft Excel file: https://1drv.ms/x/s!As32ynv0LoaIicY538vo6cxFlyYBxw?e=HKYK1T
Retrieved: 14 March 2023
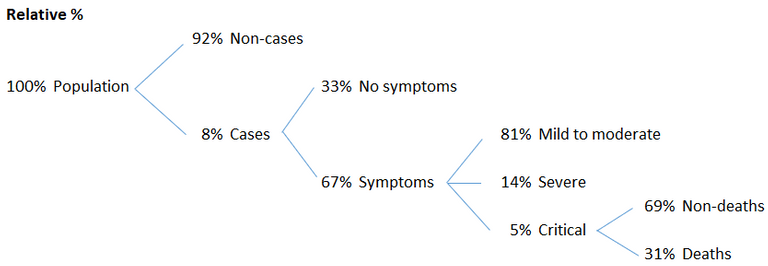
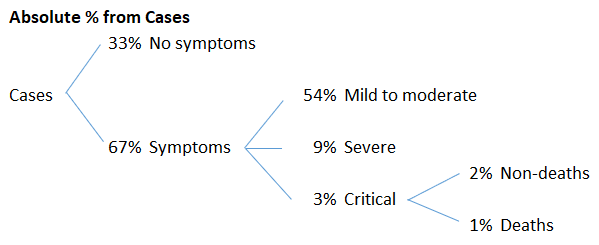
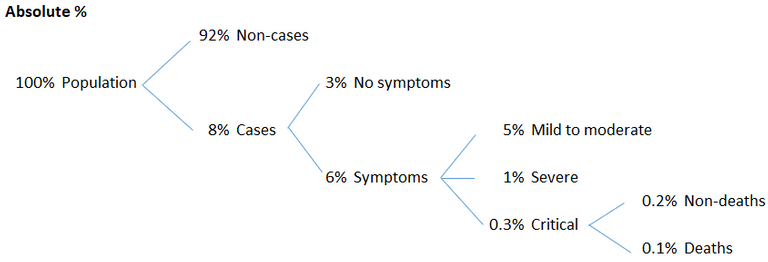

These are some big, but highly questionable numbers that need further investigation.
The Wikipedia article references the following paper on the "Patient Zero".
https://www.wsj.com/articles/in-hunt-for-covid-19-origin-patient-zero-points-to-second-wuhan-market-11614335404
Retrieved: 14 March 2023
Archive: https://archive.is/wip/yw2Cr

"His parents shopped" at a Wuhan seafood market.
The "virus" that is blamed for the above questionable statistics is the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2):
https://en.wikipedia.org/wiki/SARS-CoV-2
Retrieved: 14 March 2023
Archive: https://archive.is/wip/770NE
Colourised transmission electron micrograph of SARS-CoV-2 virions with visible coronae
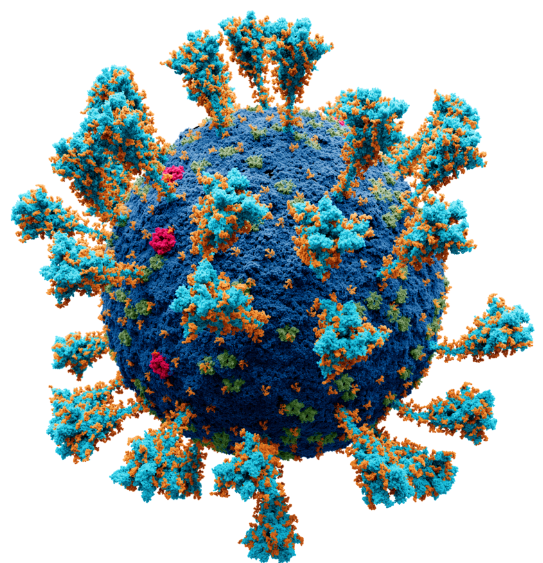
Model of the external structure of the SARS-CoV-2 virion[1]
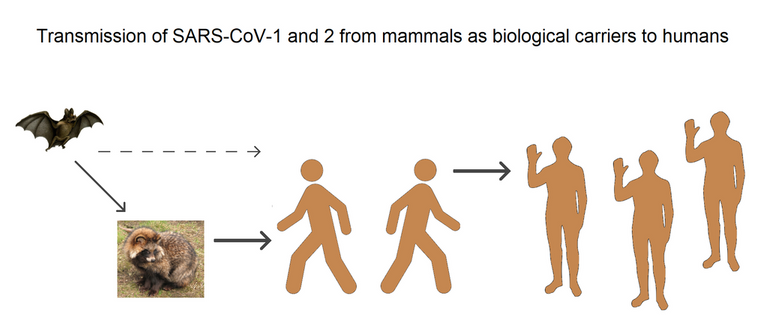
Transmission of SARS-CoV-1 and SARS‑CoV‑2 from mammals as biological carriers to humans
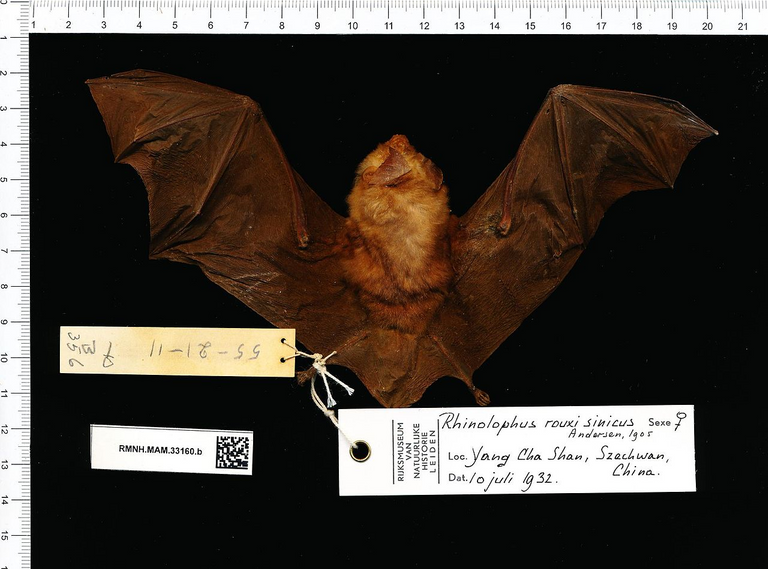
Samples taken from Rhinolophus sinicus, a species of horseshoe bats, show an 80% resemblance to SARS‑CoV‑2.
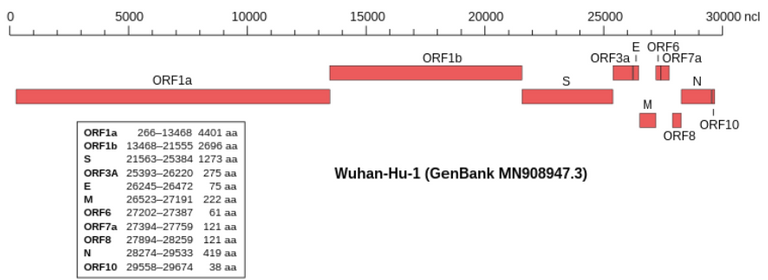
Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2
NCBI genome ID: 86693
Genome size: 29,903 bases
Year of completion: 2020
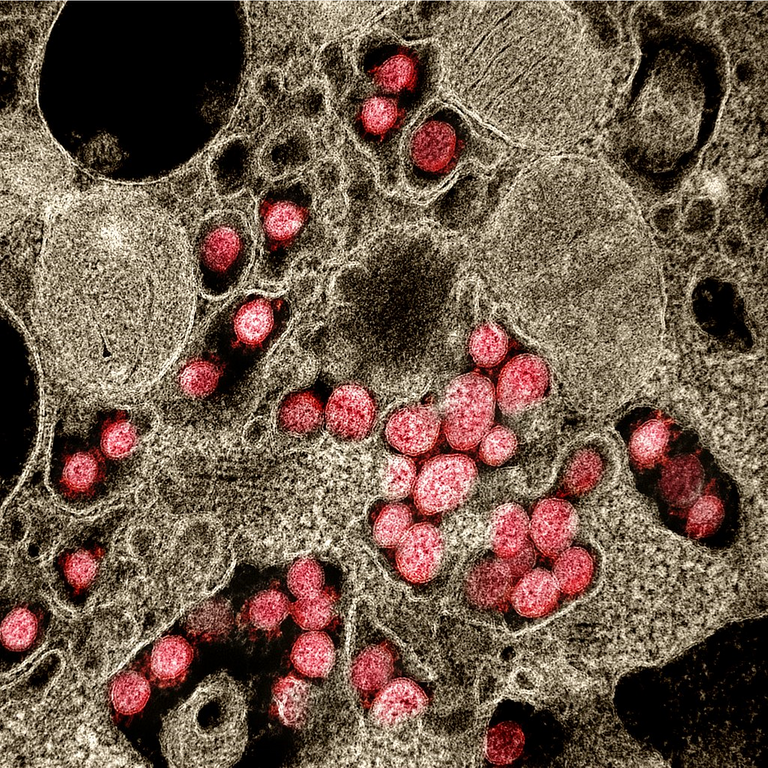
Transmission electron micrograph of SARS‑CoV‑2 virions (red) isolated from a patient during the COVID-19 pandemic
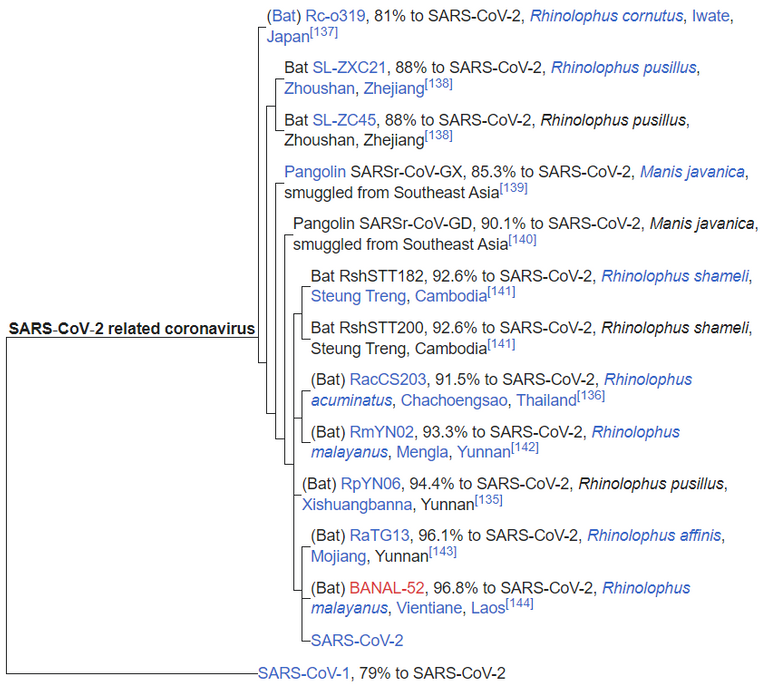
A phylogenetic tree based on whole-genome sequences of SARS-CoV-2 and related coronaviruses
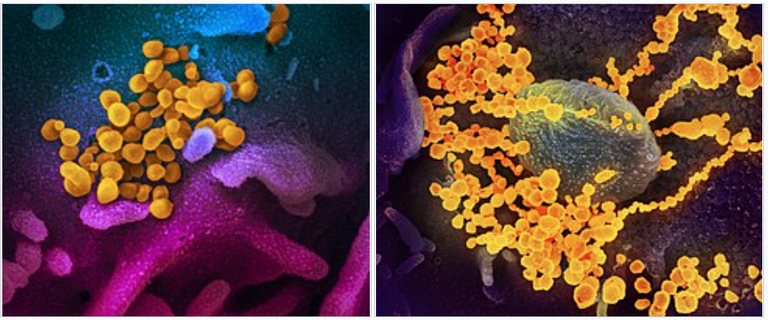
Digitally colourised scanning electron micrographs of SARS-CoV-2 virions (yellow) emerging from human cells cultured in a laboratory
ChatGPT on the Main Paper on COVID "Virus" Isolation
With the rise of Artificial Intelligence (AI), it is now quicker to gather and synthesize mainstream information.
I say "mainstream" because ChatGPT, like Wikipedia, is biased towards information that is already widely held by the mainstream institutions and public at large.
For mainstream information, I mainly cite Wikipedia, but where it comes up short, I have found that ChatGPT helps fill in the gaps.
For example, in this discussion ChatGPT gave me the main paper cited on "COVID-19 virus isolation".
https://chat.openai.com/chat
Retrieved: 18 March 2023MES: What is the main paper on isolation of the virus that causes COVID-19?
ChatGPT: The main paper on the isolation of the virus that causes COVID-19 is "A Novel Coronavirus from Patients with Pneumonia in China, 2019" by Zhang et al., published in the New England Journal of Medicine in February 2020. This paper describes the isolation and characterization of the virus from patients with severe respiratory illness in Wuhan, China, and provides key information on the genetic sequence, morphology, and pathogenicity of the virus. The findings of this paper were critical in confirming the causative agent of COVID-19 and have been widely cited in subsequent research and public health efforts related to the pandemic.
MES: give me a link of that paper
ChatGPT: Certainly! Here is the link to the paper:
Paper: A Novel Coronavirus from Patients with Pneumonia in China, 2019
The following is the paper that ChatGPT referenced as the main paper for "COVID virus isolation".
https://www.nejm.org/doi/full/10.1056/NEJMoa2001017
Retrieved: 18 March 2023
Archive: https://archive.ph/32YF4

Note that the paper was published on January 24, 2020, then updated on January 29, 2020, and appears in the February 20, 2020 issue of the New England Journal of Medicine (NEJM).
Paper Abstract
Let's go over the summary or abstract of the paper:
Summary
In December 2019, a cluster of patients with pneumonia of unknown cause was linked to a seafood wholesale market in Wuhan, China. A previously unknown betacoronavirus was discovered through the use of unbiased sequencing in samples from patients with pneumonia.
Unbiased sequencing refers to genetic sequencing techniques aimed at minimizing any biases or errors in sequencing and data processing. Pneumonia is inflammation of the lungs, primarily the alveoli, and is supposedly caused by "infection" with "viruses" or bacteria, or less commonly by other microorganisms. Inflammation is a complex biological response to harmful stimuli and is characterized by heat, pain, redness, swelling, and loss of function. Alveoli are millions of tiny air sacs in the lungs where oxygen / CO2 gas exchange occurs. Betacoronaviruses are one of four genera (classes) of coronaviruses and includes SARS-CoV-2, the alleged "virus" that "causes" COVID-19.
Continuing further in the paper:
Human airway epithelial cells were used to isolate a novel coronavirus, named 2019-nCoV, which formed a clade within the subgenus sarbecovirus, Orthocoronavirinae subfamily. Different from both MERS-CoV and SARS-CoV, 2019-nCoV is the seventh member of the family of coronaviruses that infect humans. Enhanced surveillance and further investigation are ongoing. (Funded by the National Key Research and Development Program of China and the National Major Project for Control and Prevention of Infectious Disease in China.)
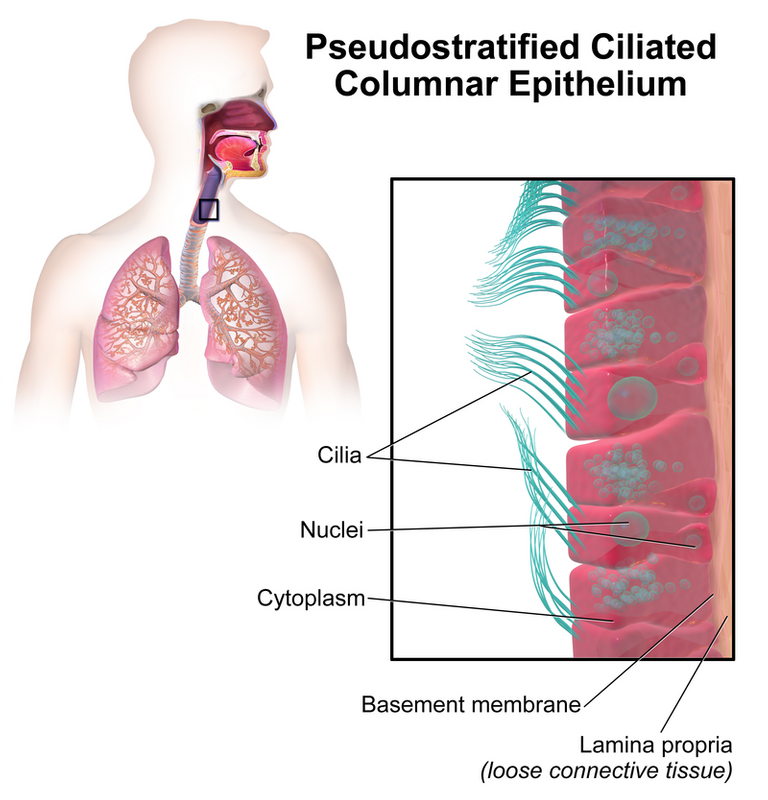
Epithelial cells are the cells that line the outer surfaces of organs and blood vessels; the human airways (nose, mouth, and down to the lungs) have the pseudostratified type. Pseudostratified is named such since they have the appearance of consisting of multiple layers of cells but are actually just one layer.
A clade is a grouping of organisms that have a common ancestor and contains all of its descendants.
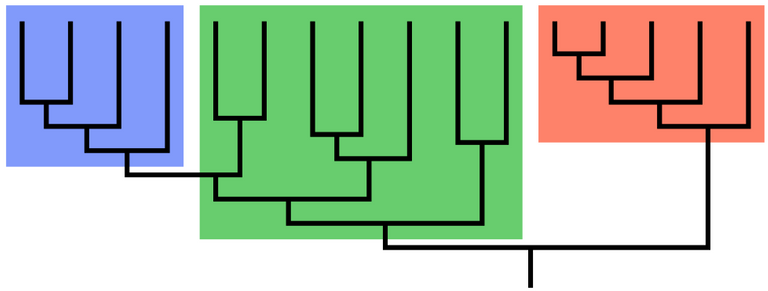
The blue and orange groupings are clades but the green is not a clade.
A subgenus is a taxonomical or grouping directly below "genus"; for example the genus Panthera has subgenera (plural of subgenus) the lion, tiger, jaguar, and leopard. Sarbecovirus (SARS Betacoronavirus) is a subgenus of Betacoronavirus. Betacoronavirus is one of four genera (plural of genus) of coronaviruses; the others are Alpha-, Gamma-, Delta-. Orthocoronavirinae is a subfamily, in the family Coronaviridae; "coronavirus" refers to any member of the Orthocoronavirinae subfamily.
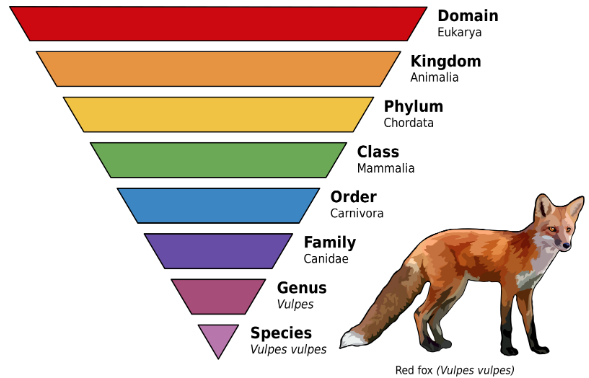
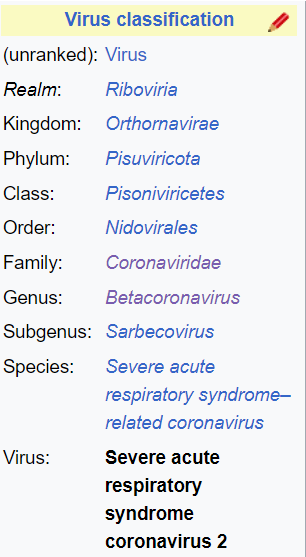
Source: Taxonomical rank for SARS-CoV 2.
Summary of the Abstract
Let's go over a quick summary of the abstract #SummaryOfTheSummary:
- Patients with pneumonia (inflammation of the lungs) of unknown cause was "linked" to a seafood market in Wuhan, China in December 2019.
- Genetic sequencing of samples from pneumonia patients was used to "discover" an unknown coronavirus.
- Human airway epithelial cells (cells that line the nose, mouth, and down to the lungs) were used to "isolate" a novel or new coronavirus, named 2019-nCoV.
Paper Introduction
Let's go over the paper's introduction:
Emerging and reemerging pathogens are global challenges for public health.1 Coronaviruses are enveloped RNA viruses that are distributed broadly among humans, other mammals, and birds and that cause respiratory, enteric, hepatic, and neurologic diseases.2,3
A pathogen is a microorganism or agent that can supposedly produce disease. A viral envelope is the outer most layer of many types of "viruses" and helps to protect the genetic material when traveling between host cells. Respiratory diseases affect the breathing organs. Enteric diseases affect the intestines or gut. Hepatic diseases affect the liver, which is responsible for detoxification, metabolism, and storage of nutrients. Neurological diseases affect the brain, spinal cord, and peripheral nerves (which extend from the brain and spinal cord to the rest of the body). Detoxification is the removal of toxic substances from the body. Metabolism is all the chemical processes that occur to maintain life; such as those involved in converting food into energy, building and repairing cells, and elimination of metabolic waste. A nerve is a cable-like bundle of fibers that transmit electrical impulses.
Continuing further in the paper.
Six coronavirus species are known to cause human disease.4 Four viruses — 229E, OC43, NL63, and HKU1 — are prevalent and typically cause common cold symptoms in immunocompetent individuals.4
Immunocompetence is the ability to have a normal "immune response" following exposure to an antigen. An antigen is any molecule or foreign matter that can bind to a specific antibody; and can trigger an "immune response". Antibodies are proteins that "identify and neutralize" foreign objects such as pathogenic bacteria and "viruses". An immune response is the body's complex and coordinated response to "invading pathogens" or foreign substances that may harm the body. The common cold is a "viral infectious disease" of the upper respiratory tract that mainly affects the respiratory mucosa (or respiratory epithelium) of the nose, throat, sinuses, and larynx. Symptoms of the common cold include: coughing, sore throat, runny nose, sneezing, headache, and fever. Fever is defined as having a body temperature above normal range, and is typically between 37.2 and 38.3°C (or 99 and 100.9°F) in humans. The upper respiratory tract is the region involved in the initial pathway for air to enter the body. The larynx or voice box is in the top of the neck involved in breathing, producing sound, and protecting the trachea from food getting inside. The sinuses are air-filled cavities in the skull connected to the nasal cavity with possible functions in moistening / filtering air and providing resonance in speech.
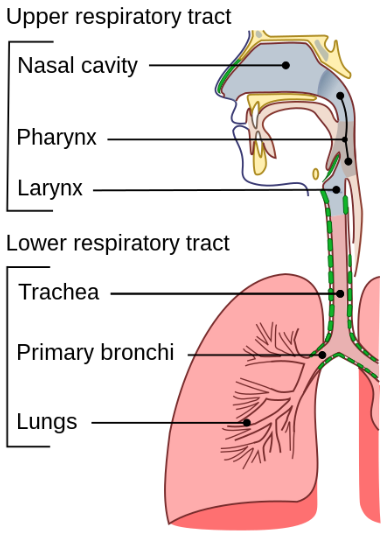
Continuing further in the paper:
The two other strains — severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) — are zoonotic in origin and have been linked to sometimes fatal illness.5
SARS-CoV (now SARS-CoV-1) is a "strain" or "variant" of a coronavirus that causes "severe acute respiratory syndrome (SARS)". Acute refers to the sudden onset and rapid progression of symptoms. Syndrome refers to a collection of symptoms associated with a disease or disorder. A zoonotic disease is an "infectious" disease caused by a pathogen that "jumps" from a non-human animal to a human, and vice versa.
Continuing further in the paper:
SARS-CoV was the causal agent of the severe acute respiratory syndrome outbreaks in 2002 and 2003 in Guangdong Province, China.6-8 MERS-CoV was the pathogen responsible for severe respiratory disease outbreaks in 2012 in the Middle East.9 Given the high prevalence and wide distribution of coronaviruses, the large genetic diversity and frequent recombination of their genomes, and increasing human–animal interface activities, novel coronaviruses are likely to emerge periodically in humans owing to frequent cross-species infections and occasional spillover events.5,10
Genetic recombination is the exchange of genetic material between different organisms resulting in the production of offspring with combinations of traits different from those found in either parent. "Viruses" recombine genomes whenever 2+ "viral" genomes are present in the same host cell. A virus is an "infectious" particle too small to be seen by optical light microscope and can only replicate inside the living cells of an organism. Spillover events occurs when a population with a high pathogen prevalence (termed a reservoir population) comes into contact with a new host population; the pathogen may or may not be transmitted within the new host. "Climate change" is said to increase the risk of "viral spillover".
Continuing further in the paper:
In late December 2019, several local health facilities reported clusters of patients with pneumonia of unknown cause that were epidemiologically linked to a seafood and wet animal wholesale market in Wuhan, Hubei Province, China.11 On December 31, 2019, the Chinese Center for Disease Control and Prevention (China CDC) dispatched a rapid response team to accompany Hubei provincial and Wuhan city health authorities and to conduct an epidemiologic and etiologic [cause of disease] investigation. We report the results of this investigation, identifying the source of the pneumonia clusters, and describe a novel coronavirus detected in patients with pneumonia whose specimens were tested by the China CDC at an early stage of the outbreak. We also describe clinical features of the pneumonia in two of these patients.
Epidemiology is the study of the distribution, patterns, risk factors, and disease conditions in a population. A wet animal wholesale market is a market that is sells live animals or animals butchered when sold; wholesale refers to selling in bulk, and typically to other businesses. Clinical features are observable or measurable characteristics associated with a disease or patient, such as symptoms and laboratory findings.
Summary of the Introduction
Let's go over the a summary of the paper's introduction:
- Supposedly there are "global challenges" of "disease-causing" microbes or microorganisms.
- Coronaviruses have an outer layer protecting RNA inside.
- Coronaviruses supposedly cause diseases relating to breathing, the gut, liver, brain, and nervous system.
- 6 coronaviruses are "known" to cause human disease.
- 4 "viruses" cause the "common cold".
- 2 other "strains" have been "linked" to fatal illness.
- SARS-CoV: "Caused" outbreaks in 2002 and 2003 in China.
- MERS-CoV: "Caused" outbreaks in 2012 in the Middle East.
- New or novel coronaviruses are "likely to emerge" in humans because coronaviruses are widespread, genetically diverse, and interactions between humans and animals are increasing.
- On December 2019, "clusters of patients" with inflammation of the lungs (aka pneumonia) of "unknown cause" were "linked" to an animal market in Wuhan, China.
- On December 31, 2019 the China CDC sent a "rapid response team" to Wuhan to investigate.
- This paper reports the results of the investigation:
- Identifies the source of the pneumonia clusters.
- Claims to detect a "novel coronavirus" in the pneumonia patients that were tested at an "early stage of the outbreak".
- Clinical features (aka symptoms and laboratory results) are presented for 2 of the pneumonia patients.
Quick Overview of Nucleic Acids and Genomes
Before I go further into the paper, let's first go over an overview of the mainstream explanations of "nucleic acids", "genomes", and cell theory in general.
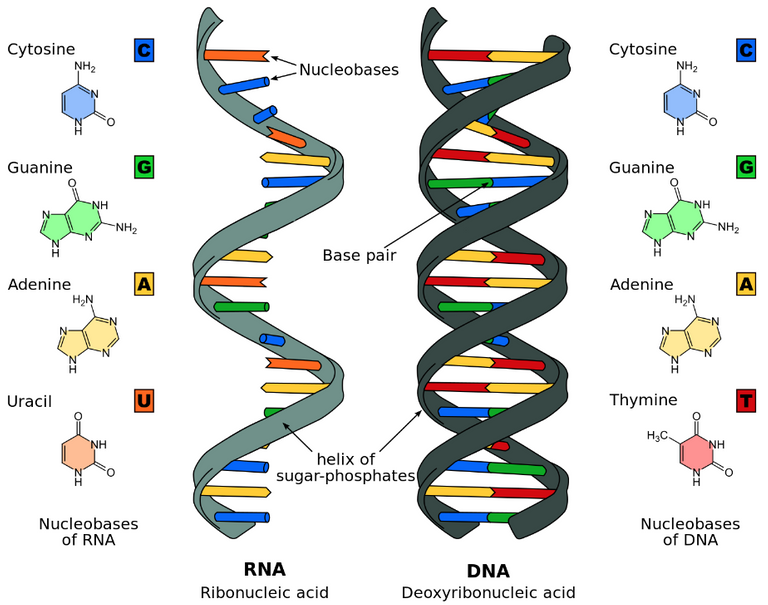
Nucleic acids are chemical compounds essential to all known forms of life. They are composed of nucleotides, which are made up of a 5-carbon sugar, a phosphate group, and a nitrogenous base (also called nucleobase). The sugar + phosphate make up the helical backbone. The specific sequence of nucleobases (CGAU in RNA, and CGAT in DNA) make up the "genetic code". Base pairs (bp) are 2 nucleobases bonded together and form the building blocks of the DNA double helix. Deoxyribonucleic acid (DNA) is made up of two polynucleotides that coil around each other to form a double helix. DNA contains genetic instructions for the development, functioning, growth, and reproduction of all known organisms and many "viruses". Ribonucleic acid (RNA) is typically a single stranded polynucleotide (and typically folded onto itself). RNA plays roles in coding, decoding, regulation, and expression of genes. Many "viruses" encode their genetic information using an RNA genome. A genome is all the genetic information of an organism and consists of the nucleotide sequences. The human genome consists of 23 pairs of chromosomes (making 46 total), including two sex chromosomes which determine the sex of the individual. Chromosomes are long DNA molecules that contain part or all of the genetic material in an organism. Cells split and duplicate the chromosomes during cell division.
Source: Typical Human cell showing the nuclear genome (inside a cell's nucleus), as well the mitochondrial genome (inside the cell's mitochondria, which is the source of the cell's energy-production).

Part of the DNA sequence of a "virus" genome.
The human genome has 3,117,275,501 base pairs; this includes the 16,569 base pairs for the human mitochondrial genome. The COVID "virus" genome has 29,903 base pairs.
Cell theory has three tenets or beliefs:
- All living organisms are composed of 1+ cells.
- The cell is the basic unit of structure and organization in organisms.
- Cells arise from pre-existing cells.
Cell theory was universally accepted but now some biologists consider non-cellular entities such as "viruses" as living organisms; which means tenet one is disputed.
The central dogma of molecular biology states that genetic information flows between DNA and RNA, and RNA to protein but not from protein to RNA.
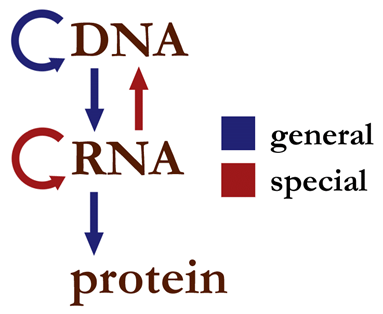
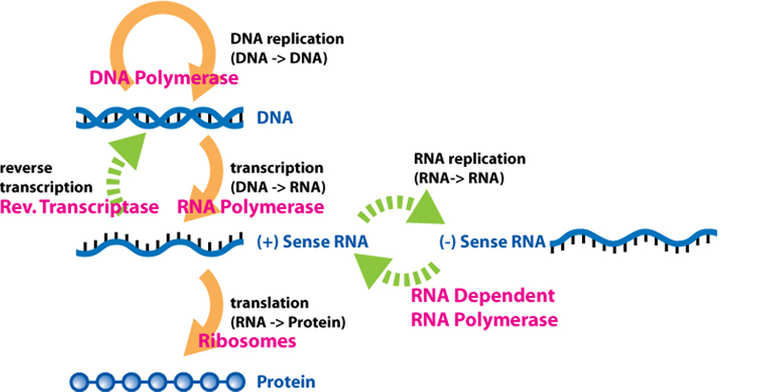
A polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. Enzymes are proteins that act as biological catalysts to accelerate chemical reactions. Polymers are very large molecules composed of many repeating subunits. Proteins are a very important type of polymer. Sense RNA refers to complementary RNA sequences; positive sense RNA can be directly translated to protein, while negative or antisense RNA can't be directly translated to protein.
Paper Methods: Detecting "Viruses"
Now let's get back to the paper and take a look at the paper's methods; the first of which involves trying to "detect known viruses".
Methods
VIRAL DIAGNOSTIC METHODS
Four lower respiratory tract samples, including bronchoalveolar-lavage fluid, were collected from patients with pneumonia of unknown cause who were identified in Wuhan on December 21, 2019, or later and who had been present at the Huanan Seafood Market close to the time of their clinical presentation. Seven bronchoalveolar-lavage fluid specimens were collected from patients in Beijing hospitals with pneumonia of known cause to serve as control samples.
The lower respiratory tract is the lower part of the groups of organs involved in moving oxygen in and out of the body. The bronchoalveolar-lavage is a method in which a tube is inserted into the lower respiratory system and fluid introduced then collected for examination.

Let's continue further into the paper.
Extraction of nucleic acids from clinical samples (including uninfected cultures that served as negative controls) was performed with a High Pure Viral Nucleic Acid Kit, as described by the manufacturer (Roche).
A cell culture is the process of growing cells under controlled conditions. A negative control is a condition expected to produce a negative result, such as giving samples without the active ingredient being tested.
Cell culture plate or "Petri dish"
Epithelial cells in culture, stained for keratin [structural protein] (red) and DNA (green)
Continuing further:
Extracted nucleic acid samples were tested for viruses and bacteria by polymerase chain reaction (PCR), using the RespiFinderSmart22kit (PathoFinder BV) and the LightCycler 480 real-time PCR system, in accordance with manufacturer instructions.12 Samples were analyzed for 22 pathogens (18 viruses and 4 bacteria) as detailed in the Supplementary Appendix.
The polymerase chain reaction (PCR) is a method of rapidly making millions to billions of complete or partial copies of a specific sample of DNA. PCR cycles refer to the repeating number of steps during DNA replication.
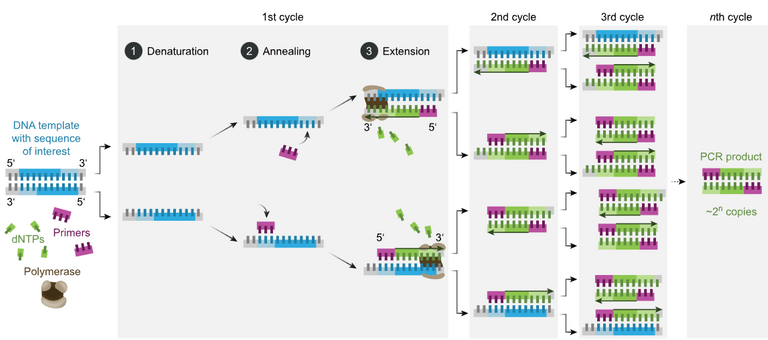
Deoxyribose nucleoside triphosphate (dNTPs) are the building blocks of DNA that are used by DNA polymerase to synthetize a new DNA strand. A Primer is a short single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis.
Continuing further:
In addition, unbiased, high-throughput sequencing, described previously,13 was used to discover microbial sequences not identifiable by the means described above. A real-time reverse transcription PCR (RT-PCR) assay was used to detect viral RNA by targeting a consensus RdRp region of pan β-CoV, as described in the Supplementary Appendix.
High-throughput sequencing or second-generation sequencing or next-generation sequencing (NGS) refers to advancements in sequencing technology that allows for entire genomes to be sequenced at once. The term unbiased refers to the method where specific sequences are not preferentially amplified or sequenced over others.
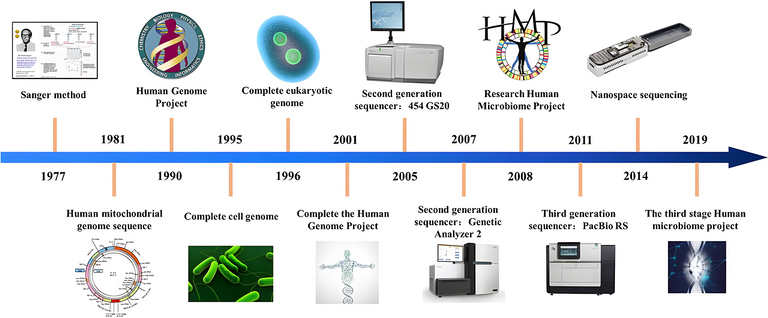
History of sequencing technology[50]
Real-time polymerase chain reaction (real-time PCR, or qPCR) monitors the replication of a targeted DNA molecule during the PCR in real time, and not at its end, as in conventional PCR. Reverse transcription PCR (RT-PCR) combines reverse transcription of RNA into DNA and then replication of the DNA targets using PCR. Reverse transcription is the process in which the enzyme reverse transcriptase is used to generate DNA from an RNA template. An assay is an investigative procedure to detect or measure a specific biological entity or function.
Pan β-CoV refers to a group or test designed to detect a broad range of betacoronaviruses. RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. The RdRp region refers to a specific region of the genome of RNA "viruses" that encodes for the RdRp enzyme. The consensus RdRp region refers to a segment of the RdRp gene that is highly conserved among different "strains" or "isolates" of a "virus species".
Summary of Detecting the "Virus"
Let's quickly summarize the "virus detection" methodology in this paper.
- 4 lower respiratory fluid samples were taken from patients with "pneumonia of unknown cause" in Wuhan on December 21, 2019 or later.
- 7 lower respiratory fluid samples were taken from patients with "pneumonia of known cause" in Beijing to serve as control samples.
- Nucleic acids were extracted from the clinical samples and "uninfected" cultures.
- PCR (or DNA replication) tests were done on the extracted nucleic acids to test for 22 "viruses" and 4 bacteria.
- High-throughput sequencing were used to discover microbial sequences not identifiable by the above means.
• RT-PCR (RNA conversion to DNA then DNA replication) assay was used to detect "viral RNA" by targeting a consensus RdRp region of a group of betacoronaviruses, the genera of coronaviruses that includes the alleged COVID-19 "virus" titled SARS-CoV-2.
Paper Methods: Isolating the "Virus"
Now let's take a look at the next section in the above paper, which is on allegedly isolating the "virus".
ISOLATION OF VIRUS
Bronchoalveolar-lavage fluid samples were collected in sterile cups to which virus transport medium was added.
Viral transport medium (VTM) is a chemical preservative used to preserve "virus" specimens during transport instead of using cumbersome super-cooling equipment. Typical VTM components may include saline solution, phosphate-buffered saline (PBS), or fetal bovine serum (FBS). A buffer solution is an acid or base aqueous solution used to maintain a constant pH when a small amount of strong acid or base is added to it. Acids and bases are defined as in the 3 theories shown in the image below. The pH or "potential or power of hydrogen" indicates the acidity or basicity of an aqueous solution; the higher the value the more basic; the lower the more acidic. An aqueous solution is a solution in which the solvent is water. A solvent is a substance that dissolves another substance, called a solute, resulting in a solution. Fetal bovine serum (FBS) is extracted from blood drawn from a bovine (cow, bull, or oxen) fetus after it is killed at a slaughterhouse. Serum is the part of the blood not involved in blood clotting.
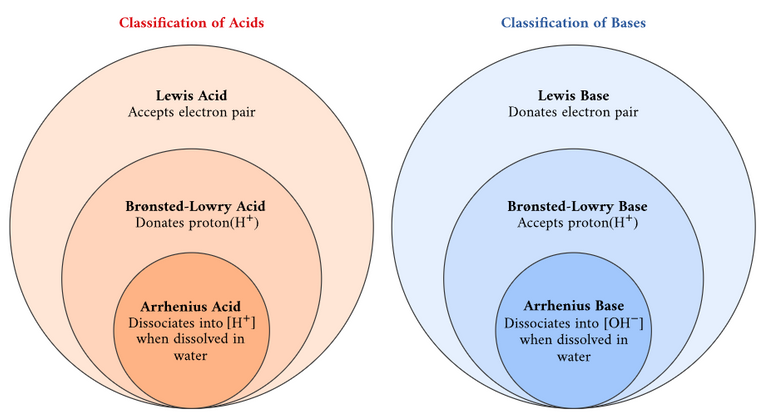
Classification of acids and bases.
Continuing further in the paper:
Samples were then centrifuged to remove cellular debris. The supernatant was inoculated on human airway epithelial cells,13 which had been obtained from airway specimens resected from patients undergoing surgery for lung cancer and were confirmed to be special-pathogen-free by NGS [Next-Generation Sequencing].14
Centrifugation involves rapid spinning to separate objects by density. The supernatant or supernate is the remaining liquid after centrifugation or settling. Inoculation is the introducing of microbes into the sample. Resection is the removal of all or a key part of an internal organ or body part. Cancer refers to abnormal cell growth with the potential to "invade" or "spread" in the body. Specific-pathogen-free (SPF) refers to being free of specific "pathogens".
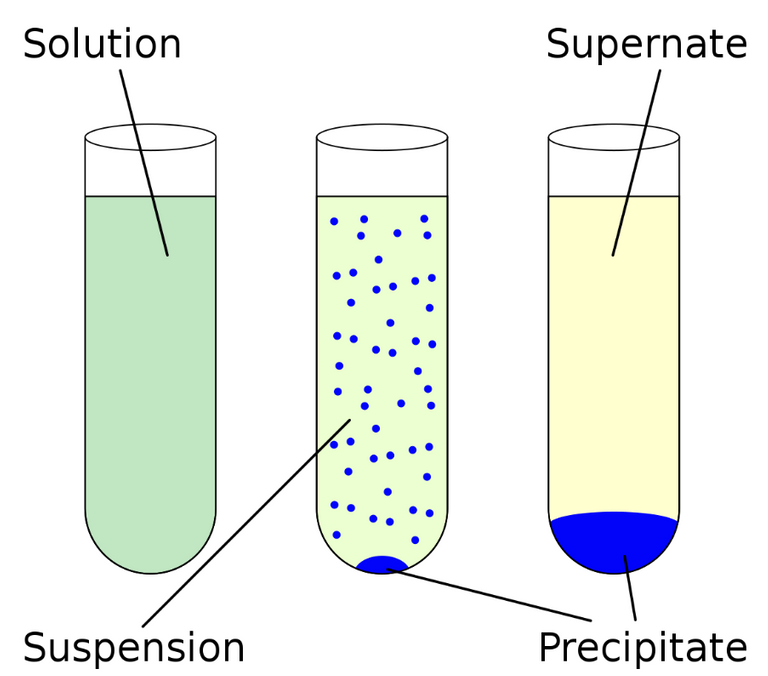
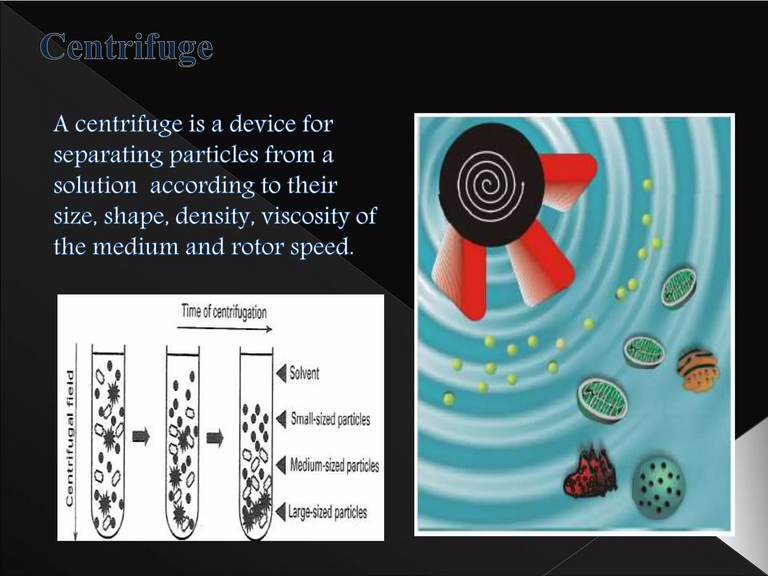
Schematic of centrifugation
Continuing further in the paper:
Human airway epithelial cells were expanded on plastic substrate to generate passage-1 cells and were subsequently plated at a density of 2.5×105 cells per well on permeable Transwell-COL (12-mm diameter) supports.
Plastic substrate refers to a plastic surface on which cells are grown. Expanding cells means growing cells outside of their natural environment. Passage-1 cells refer to cells grown and expanded in culture for the first time after obtaining from a primary cell source such as a tissue or organ. Plating refers to growing cells on dishes or multiwell plates which are used to produce more consistent and reproducible population of cells.

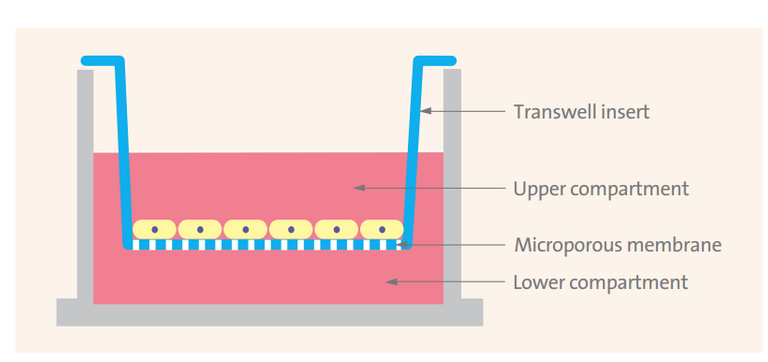
Schematic of a Transwell insert
Continuing further in the paper:
Human airway epithelial cell cultures were generated in an air–liquid interface for 4 to 6 weeks to form well-differentiated, polarized cultures resembling in vivo pseudostratified mucociliary epithelium.13
Air liquid interface cell culture (ALI) involves growing cells in which the bottom portion are in contact with a liquid medium and the top exposed to air; this is to better mimic in vivo conditions. In vivo (Latin for "within the living") refers to tests done on the natural living organisms; in contrast in vitro (Latin for "within the glass") refers to tests done in a laboratory setting outside of normal biological conditions. Cellular differentiation is the process in which one cell changes from one type to another; stem cells can differentiate into various cell types and the further that cells differentiate the more specific type of cell they become. Cell polarity refers to spatial differences in shape, structure, and function within a cell. Mucociliary refers to the combination of mucus and cilia in the respiratory system; mucus is a sticky substance that helps trap foreign particles and cilia are hair-like structures on the surface of epithelial cells and beat in coordinated manner to move the mucus and trapped particles up and out of the respiratory tract.
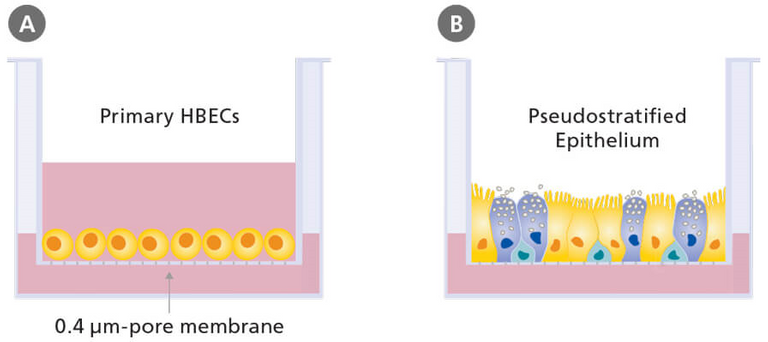
Schematic of submerged (A) and air-liquid interface (B) culture of primary human bronchial epithelial cells (HBEC) grown using porous culture inserts.
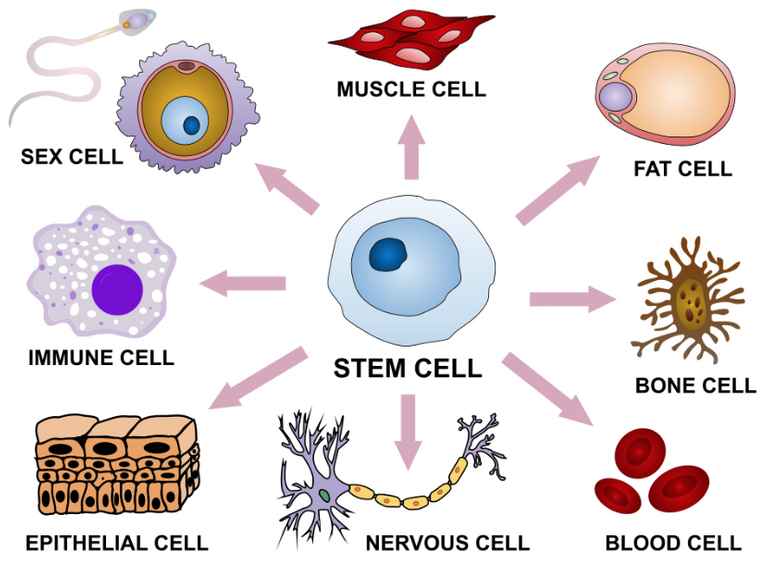
Stem cell differentiation into various tissue types.
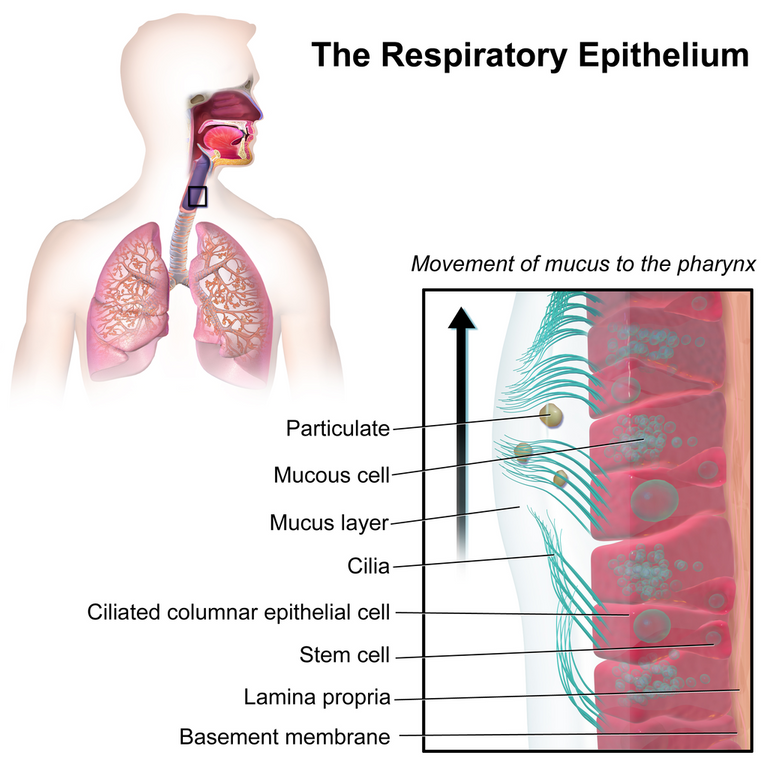
Illustration depicting the respiratory epithelium.
Continuing further in the paper:
Prior to infection, apical surfaces of the human airway epithelial cells were washed three times with phosphate-buffered saline; 150 μl of supernatant from bronchoalveolar-lavage fluid samples was inoculated onto the apical surface of the cell cultures.
Apical refers to the top part of the cells which extend into the open airway. A microlitre or μL is 10-6 L, which is equivalent to a cubic millimeter or mm3. 150 μL = 0.15 mL.
MES Note: How does the paper know there is a "virus" in which they are "infecting" the cell cultures with?
Continuing further in the paper:
After a 2-hour incubation at 37°C, unbound virus was removed by washing with 500 μl of phosphate-buffered saline for 10 minutes; human airway epithelial cells were maintained in an air–liquid interface incubated at 37°C with 5% carbon dioxide. Every 48 hours, 150 μl of phosphate-buffered saline was applied to the apical surfaces of the human airway epithelial cells, and after 10 minutes of incubation at 37°C the samples were harvested. Pseudostratified mucociliary epithelium cells were maintained in this environment; apical samples were passaged in a 1:3 diluted vial stock to new cells.
An incubator maintains optimal temperature, humidity, and other conditions to grow and maintain cell cultures; 37°C is typically chosen because it approximates the normal body temperature of humans. Serial passage is the process of growing bacteria or a "virus" in iterations: by growing in one environment then removing a portion of the population and putting into a new environment. "1:3 diluted vial stock to new cells" means 1 part of the original sample was mixed with 3 parts of the new culture medium.
Continuing further in the paper:
The cells were monitored daily with light microscopy, for cytopathic effects, and with RT-PCR, for the presence of viral nucleic acid in the supernatant. After three passages, apical samples and human airway epithelial cells were prepared for transmission electron microscopy.
Light microscopy or optical microscopy refers to microscopes that use visible light and a system of lenses to magnify images. Cytopathic effect or cytopathogenic effect or CPE refers to structural changes in host cells "caused by viral invasion"; such as lysis which is the breaking down of the cell membrane, or such as cell death without lysis due to an inability to replicate.
Summary of Isolating the "Virus"
Let's quickly summarize the "virus isolation" methodology used in this paper.
- Human airway fluid samples were collected from patients and mixed with a chemical preservative (or fetal bovine serum).
- The samples were spun really fast (in a centrifuge) to remove cellular debris.
- The supernatant was then removed and used to "infect" human airway epithelial cells obtained from lung cancer surgery patients and grown as a cell culture on a plastic surface.
- Prior to "supernatant infection", the top part of the human airway epithelial cells was washed 3 times with a water-based salt solution (PBS).
- 0.15 mL of supernatant was used in the "infection" and was placed on the apical surfaces (top surfaces) of the cell cultures.
- After 2-hours of incubation at 37°C (normal human body temperature) with 5% CO2 in an air-liquid interface, "unbound virus" was removed by washing with 0.50 mL of PBS for 10 minutes.
- Every 48 hours, 0.15 mL of PBS was applied to the apical surfaces of the cell cultures, then "incubated" at 37°C for 10 minutes, then harvested (i.e. some apical surfaces of the cell cultures were removed from the culture).
- Pseudostratified mucociliary epithelium cells were maintained in the above environment.
- The removed/harvested apical samples were diluted at a ratio of 1 to 3 to a new cell culture.
- The new cell culture was monitored daily with an optical microscope for cell breakdown or cell death (aka "cytopathic effects").
- The supernatant of the new cell culture was monitored with RT-PCR for the presence of "viral nucleic acid".
- The above procedure was done again 2 more times (aka 3 total "passages") starting from the supernatant at the last step above then "infecting" a new set of human airway epithelial cells.
- After these 3 total passages, the apical samples (supernatant) and (bottom) human airway epithelial cells were prepared for Transmission Electron Microscopy (TEM).
Paper Methods: Electron Microscope Preparation
Now let's take a look at the next part of the paper, which is preparing the samples for electron microscopy.
TRANSMISSION ELECTRON MICROSCOPY
Supernatant from human airway epithelial cell cultures that showed cytopathic effects was collected, inactivated with 2% paraformaldehyde for at least 2 hours, and ultracentrifuged to sediment virus particles.
Virus inactivation refers to the process of removing "viruses" completely or rendering them "non-infectious". Paraformaldehyde is a chemical preservative used to "fix" proteins or other molecules in the sample via cross-linking polymers, thus locking them in place and preventing any further changes. Fixation is the preservation of biological tissues from decay or degradation, to make them suitable for subsequent analysis. A cross-link is a bond or a short sequence of bonds that links one polymer chain to another. The 2% Paraformaldehyde indicates the concentration of the solution: 2 grams of paraformaldehyde for every 100 mL of solution. An ultracentrifuge is a very fast centrifuge, with speeds about 100,000 RPM (Revolutions Per Minute). Transmission electron microscopy (TEM) involves firing a beam of electrons through an ultrathin section and the resulting interactions with the sample forms an image that can be made visible through various methods, such as a fluorescent screen. Electron microscopes provide far greater zoom than optical microscopes but the samples used are of dead material because of the high energy electron beam and typically stained with heavy metals. Heavy metals have relatively high densities or atomic masses. Fluorescence is the emission of light by a substance that has absorbed electromagnetic radiation. Electrons and electromagnetic radiation (EMR) are not properly defined by mainstream science but a good way to view them is as interactions with an all pervasive medium, the Aether; electrons can be viewed as vibrations in the Aether and EMR as the resulting waves in the Aether. Some good resources on this are Ionel Dinu and Fractal Woman.
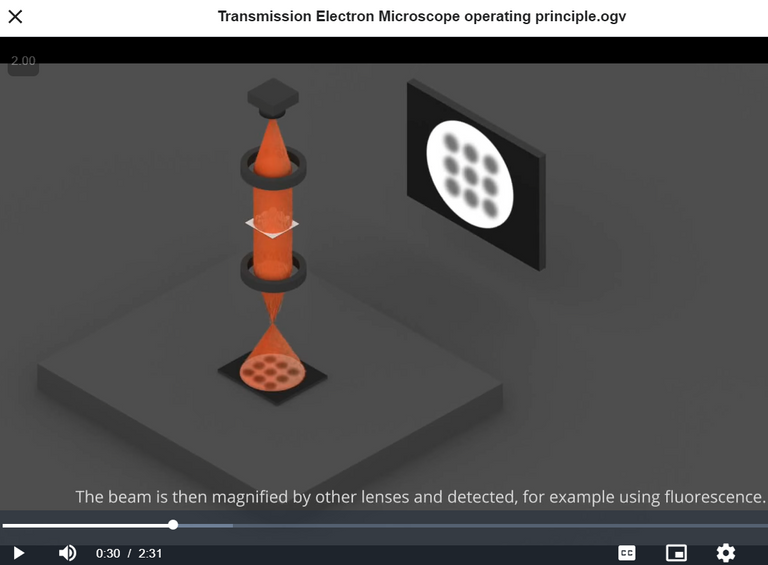
Continuing further in the paper:
The enriched supernatant was negatively stained on film-coated grids for examination.
Enriched supernatant refers to the portion of the centrifuged supernatant that contains the sedimented "virus" particles; the term "enriched" refers to the concentration of the solute (in this case the "virus" particles) has increased. A negative stain involves staining the background while the sample is not stained; in contrast, positive staining involves staining the actual sample. Staining is a technique to increase contrast in samples; in electron microscopy electron-dense compounds of heavy metals are typically used. Contrast is the degree of differences in color that makes an object visibly distinguishable. Film-coated grids are a type of electron microscopy sample grid that has a thin layer coated onto its surface to provide a hydrophilic surface for the sample to adhere to; while the grids themselves are typically made of a thin metal mesh such as copper or nickel. Hydrophilic (Greek for "water-loving") refers to molecules that are attracted to water and tend to be dissolved by water.
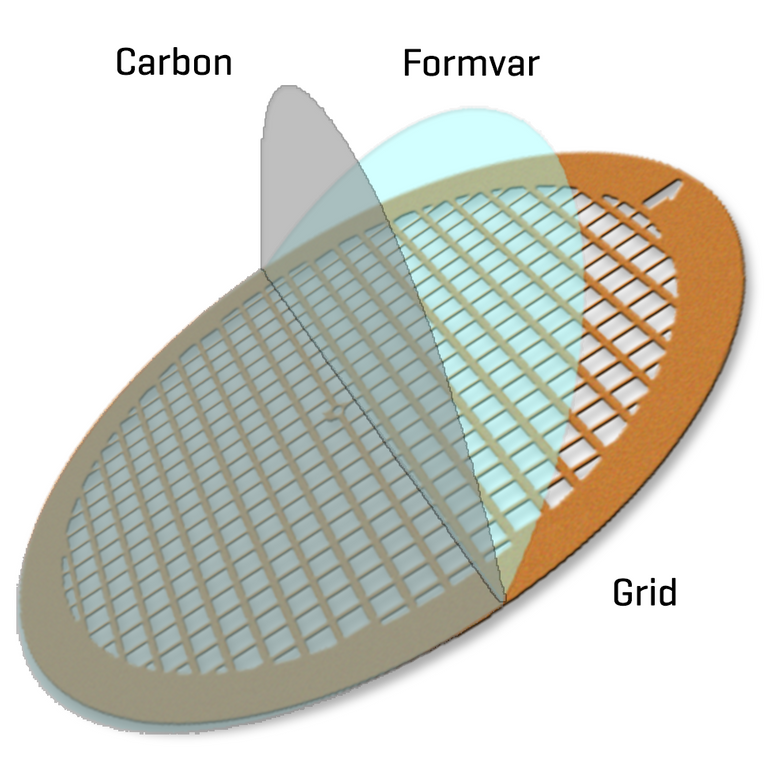
Formvar polymer + carbon coated grid
Continuing further in the paper:
Human airway epithelial cells showing cytopathic effects were collected and fixed with 2% paraformaldehyde–2.5% glutaraldehyde and were then fixed with 1% osmium tetroxide dehydrated with grade ethanol embedded with PON812 resin.
Glutaraldehyde is another crosslinking chemical compound. Osmium tetroxide is a heavy metal chemical compound used for fixation and/or staining. Ethanol is a type of alcohol. Grade ethanol refers to highly purified ethanol, free of impurities that could affect the experimental results. Ethanol is a dehydrating agent that is able to penetrate cell membranes and replace water molecules within the cells. PON812 resin is an industrial resin. A resin is a solid or highly viscous substance that is typically convertible into polymers. Viscosity is a measure of a fluid's resistance to deformation; for example syrup has a higher viscosity than water. Embedding samples in a solid support material, such as a resin, is to provide structural support for the sample during further processing and imaging.
Continuing further in the paper:
Sections (80 nm) were cut from resin block and stained with uranyl acetate and lead citrate, separately. The negative stained grids and ultrathin sections were observed under transmission electron microscopy.
1 nm = 10-9m = 0.001 um. Uranyl acetate is a heavy metal stain derived from uranium. Lead citrate is a heavy metal stain derived from Lead.
Summary of Electron Microscope Preparation
Let's quickly summarize the TEM sample preparation methodology in this paper.
- Supernatant from the "infected" human airway cell cultures that showed "cytopathic effects" (aka cell membrane breakdown or cell death) was collected.
- The supernatant was "inactivated" with the chemical preservative paraformaldehyde that "fixes" or locks biomolecules in place by "cross-linking" or attaching to polymers.
- After 2+ hours of "inactivation", the supernatant was ultracentrifuged, which involves spinning at really fast speeds of about 100,000 RPM, to sediment "virus" particles.
- The portion of the supernatant that contains the sedimented "virus" particles, hence called "enriched supernatant", was placed on thin film-coated metal grids that were stained with heavy metals (i.e. negative staining).
- Human airway cells that showed cytopathic effects were collected and fixed with more chemical preservatives, fixed with a heavy metal preservative, dehydrated with alcohol, and embedded in a resin block to provide structural support.
- Ultrathin sections of 80 nm were cut from the resin block and stained with 2 heavy metal stains in separate steps.
- The centrifuged supernatant on the negative stained grids and the human airway cells on the ultrathin resin block sections were observed under electron microscopy.
Paper Methods: Sequencing the "Virus"
So far, none of the steps covered even remotely show any common sense demonstrations of "isolation", let alone what the paper is trying to achieve.
Let's now take a look at the next section of the paper, which is on sequencing the "virus" from the cell cultures.
Note that I will not go over too many of the definitions of the specific industry methods for DNA sequencing mentioned in the paper.
RNA extracted from bronchoalveolar-lavage fluid and culture supernatants was used as a template to clone and sequence the genome. We used a combination of Illumina sequencing and nanopore sequencing to characterize the virus genome.
Illumina sequencing involves generating millions of short sequence reads, about 150-300 base pairs in length), in parallel. Nanopore sequencing is a new technology that can generate very long continuous sequence reads, several thousand bases in length. The short reads in Illumina sequencing helped correct errors in the long continuous reads in nanopore sequencing.
Continuing further in the paper:
Sequence reads were assembled into contig maps (a set of overlapping DNA segments) with the use of CLC Genomics software, version 4.6.1 (CLC Bio).
A contig (from contiguous) is a continuous sequence of DNA that has been assembled from a set of overlapping DNA fragments, called reads. Contiguous means objects are physically close together. A contig map is a graphical representation of the physical organization of contigs in a genome or chromosome.
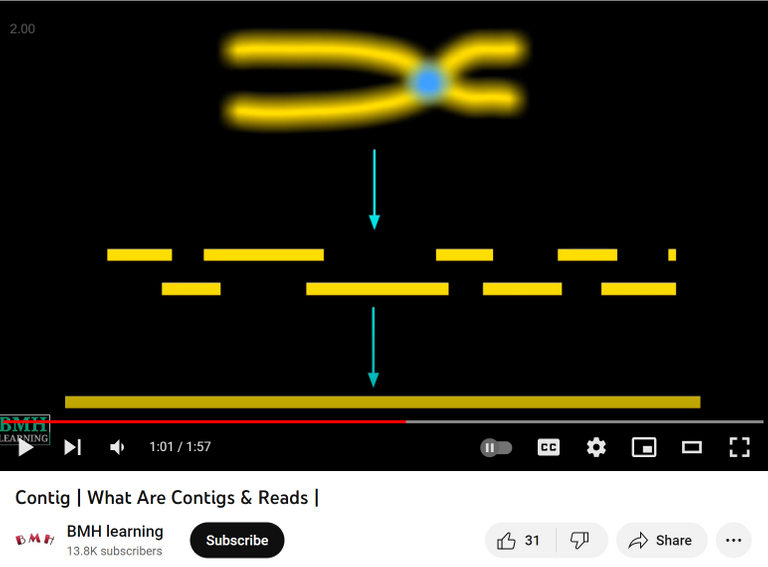
Video tutorial: Contigs are continuous sequences formed from overlapping sequences.
Continuing further in the paper:
Specific primers were subsequently designed for PCR, and 5′- or 3′-RACE (rapid amplification of cDNA ends) was used to fill genome gaps from conventional Sanger sequencing. These PCR products were purified from gels and sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit and a 3130XL Genetic Analyzer, in accordance with the manufacturers’ instructions.
A Primer is a short single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. The 5' or 3' refer to orientation of a single strand of nucleic acid. Complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA template by the RT enzyme. Sanger sequencing or chain-termination sequencing involves replicating DNA using modified nucleotides (ddNTPs) to terminate the DNA (cDNA in this case) chain elongation and allows for generating a series of DNA fragments that differ in length by a single nucleotide.
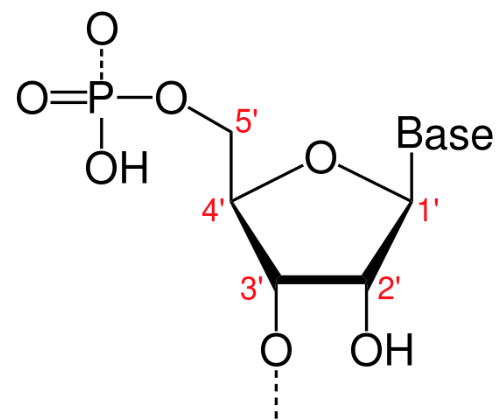
A furanose (sugar-ring) molecule with carbon atoms labeled using standard notation. The 5′ is upstream; the 3′ is downstream. DNA and RNA are synthesized in the 5′-to-3′ direction.
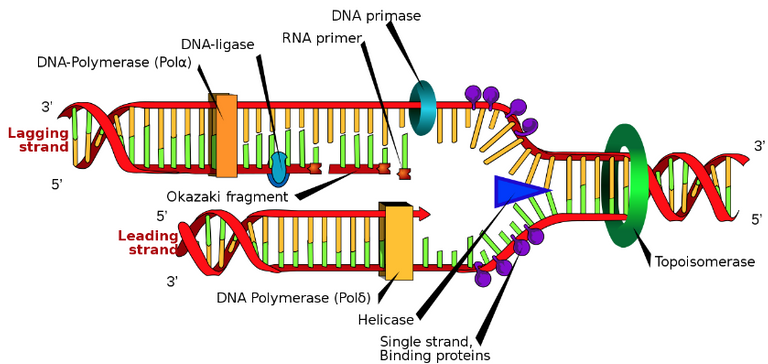
Schematic: The DNA replication fork. RNA primer labeled at top. Video animation: The fork splits the DNA double strand into 2 single strands, and the leading strand replicates continuously while the lagging strand replicates discontinuously.
Continuing further in the paper:
Multiple-sequence alignment of the 2019-nCoV and reference sequences was performed with the use of Muscle. Phylogenetic analysis of the complete genomes was performed with RAxML (13) with 1000 bootstrap replicates and a general time-reversible model used as the nucleotide substitution model.
Multiple-sequence alignment is the process of aligning the genomic sequences to identify similarities or differences. Phylogenetics is the study of the evolutionary history and relationships among or within groups of organisms. Bootstrap replicates are a statistical technique to estimate the probability of a phylogenetic tree by generating multiple random samples of the original data set. Substitution model is a model describing the changes in DNA sequences over "evolutionary time"; a time reversible model does not care which sequence is the ancestor and which is the descendant.
Summary of Sequencing the "Virus"
Here is a quick summary of the "virus" sequencing procedure in this paper:
- RNA from patient airway fluid + cell culture supernatants were extracted and assumed to be from a "virus".
- The genome of this RNA-assumed "virus" was produced using a combination of Illumina sequencing (which produces millions of short sequences) and nanopore sequencing (which produces continuous long sequences).
- The short reads are used to correct errors in the long reads.
- Primers, or short nucleic acids that initiate DNA synthesis, were designed for PCR tests.
- DNA fragments from PCR were then sequenced.
- "Virus" sequences were compared with reference sequences.
- Statistical analysis was performed on the likely "evolutionary history" of the "virus" genome.
The question still remains, where is this "virus"? How do they know there is a "virus"? Why is the paper throwing the word "virus" around so casually? #Questions
RNA Assumed to be from a "Virus"
After spending hours combing through the paper with a fine-tooth comb trying to comprehend where or what this "virus" is, I finally realized that the authors were assuming that RNA detected in the samples were from a "virus". Here I expand further on this assumption.
Recall the earlier statement by the paper:
RNA extracted from bronchoalveolar-lavage fluid and culture supernatants was used as a template to clone and sequence the genome. We used a combination of Illumina sequencing and nanopore sequencing to characterize the virus genome.
Let's ask #ChatGPT to explain what this means exactly.
https://chat.openai.com/chat
Retrieved: 8 May 2023MES: what does this mean:
RNA extracted from bronchoalveolar-lavage fluid and culture supernatants was used as a template to clone and sequence the genome. We used a combination of Illumina sequencing and nanopore sequencing to characterize the virus genome.
ChatGPT: This sentence describes a molecular biology experiment that involved isolating RNA from two sources (bronchoalveolar-lavage fluid and culture supernatants) and using it as a template to clone and sequence the genome of a virus.
…
MES: how do you know the RNA is from a virus?
ChatGPT: In the sentence you provided, the assumption is that the RNA extracted from bronchoalveolar-lavage fluid and culture supernatants contains viral RNA. This assumption is based on prior knowledge or suspicion that the samples contain a viral infection, or that the researchers were specifically looking for viral RNA.
In general, the presence of viral RNA in a biological sample can be confirmed through several methods, including:
PCR-based assays: These assays use specific primers that are designed to amplify regions of viral RNA or DNA, allowing for the detection of viral genetic material in the sample.
Sequencing: Once viral RNA has been detected and amplified, the sequence of the RNA can be determined and compared to known viral sequences in databases, allowing for the identification of the specific virus.
Immunological assays: These assays use antibodies that are specific to viral proteins or antigens, allowing for the detection and identification of the virus in the sample.
In the context of the sentence you provided, it is likely that the researchers used one or more of these methods to confirm the presence of viral RNA in the bronchoalveolar-lavage fluid and culture supernatants before proceeding with the cloning and sequencing of the virus genome.
Summary of RNA "Virus" Assumption
Let's quickly summarize the #ChatGPT explanations for the assumption of RNA being from "viral origin".
- RNA obtained from patient samples or cell cultures may be assumed to be from a "virus" if:
- Observations that the sample contain a "viral infection", such as from observing "cytopathic effects".
- "Viral RNA" is specifically being searched for.
- Several methods can be used to confirm the presence of "viral RNA":
- PCR-amplification of specific regions of previously determined "viral RNA or DNA".
- Sequenced "viral RNA" can be compared with known "viral" sequences in databases.
- Use antibodies "specific to viral proteins or antigens" to identify the "virus" in the sample.
These are some interesting concepts, which will need to be explored in more detail in a future video.
Nonetheless, it still remains highly questionable that a bunch sequences of nucleotides (aka RNA) are somehow assumed to be "disease causing particles that can infect and replicate through a host and spread to other individuals".
Paper Results
Now let's go over the results of the paper.
Paper Results: Patients
The first part of the results went over the patients used in the paper / research.
Patients
Three adult patients presented with severe pneumonia and were admitted to a hospital in Wuhan on December 27, 2019. Patient 1 was a 49-year-old woman, Patient 2 was a 61-year-old man, and Patient 3 was a 32-year-old man. Clinical profiles were available for Patients 1 and 2. Patient 1 reported having no underlying chronic medical conditions but reported fever (temperature, 37°C to 38°C) and cough with chest discomfort on December 23, 2019. Four days after the onset of illness, her cough and chest discomfort worsened, but the fever was reduced; a diagnosis of pneumonia was based on computed tomographic (CT) scan. Her occupation was retailer in the seafood wholesale market.
A clinical profile is a detailed summary of a patient's medical history. Tomography is imaging by sections using a kind of penetrating wave. A computed tomography (CT) scan uses X-rays to penetrate through the body and measure the loss of intensity through the body. Radiography is the general imaging technique using X-rays, gamma rays, or similar radiation to view the internal form of an object.
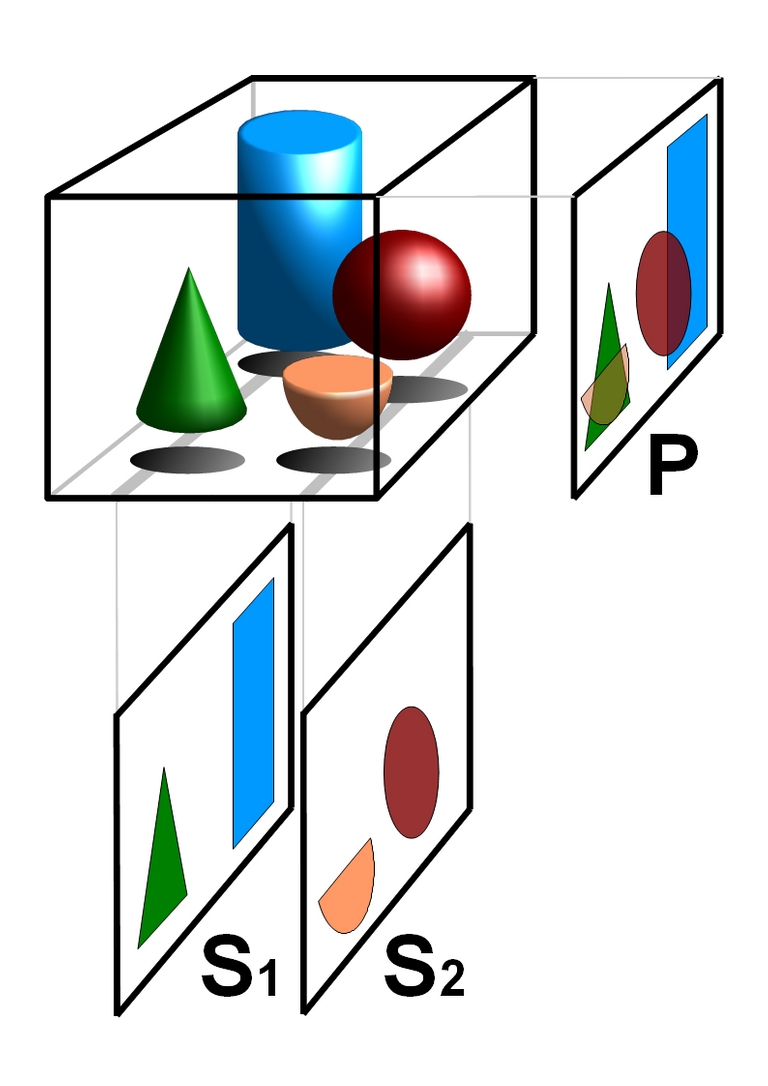
Basic principle of tomography: superposition free tomographic cross sections S1 and S2 compared with the (not tomographic) projected image P
CT scanner
Continuing further into the paper:
Patient 2 initially reported fever and cough on December 20, 2019; respiratory distress developed 7 days after the onset of illness and worsened over the next 2 days (see chest radiographs, Figure 1), at which time mechanical ventilation was started. He had been a frequent visitor to the seafood wholesale market. Patients 1 and 3 recovered and were discharged from the hospital on January 16, 2020. Patient 2 died on January 9, 2020. No biopsy specimens were obtained.
Respiratory distress is the condition of having trouble breathing. Mechanical ventilation is the use of a ventilator to provide artificial ventilation, which is the moving of air into and out of the lungs. Discharged refers to releasing a patient from a healthcare facility after receiving medical treatment. A biospy is the removing of a sample of tissue from a body for examination to determine the presence or extent of a disease.
Ventilator
Continuing further in the paper:
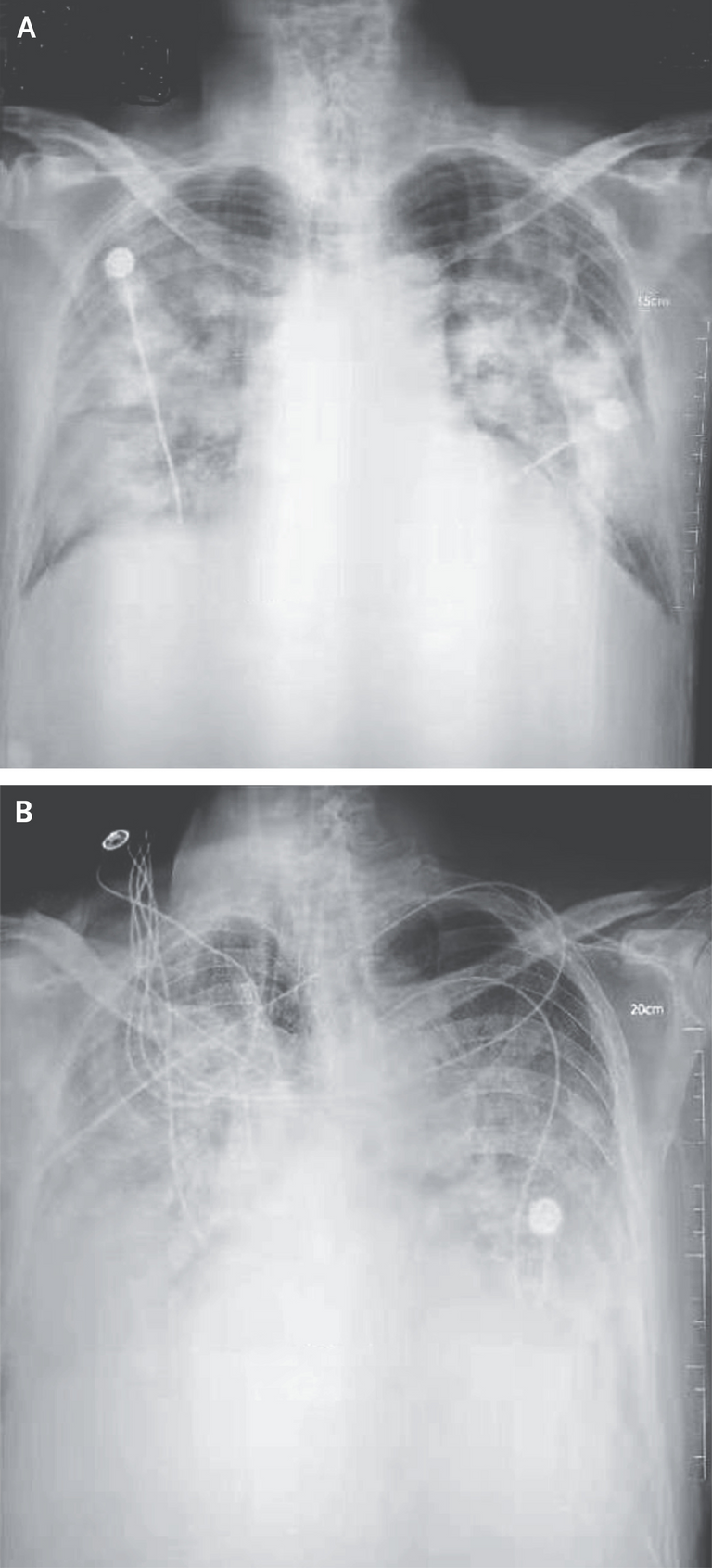
Figure 1. Chest Radiographs.
Shown are chest radiographs from Patient 2 on days 8 and 11 after the onset of illness. The trachea was intubated and mechanical ventilation instituted in the period between the acquisition of the two images. Bilateral fluffy opacities are present in both images but are increased in density, profusion, and confluence in the second image; these changes are most marked in the lower lung fields. Changes consistent with the accumulation of pleural liquid are also visible in the second image.
The trachea or windpipe is the tube that connects the larynx to the lungs. Intubation is the medical procedure involving the insertion of a tube into the body. Bilateral fluffy opacities refer to radiological findings on chest images involving cloudy or hazy areas on both sides of the lungs, indicating various medical conditions. Opacity is the measure of impenetrability of electromagnetic radiation, or other kinds of radiation. Profusion refers to the extent or degree of something. Confluence refers to the merging of multiple structures, in this case fluffy opacities. The pleurae are two opposing layers overlying the lungs and inside the chest wall. Pleural liquid is the fluid inside the space between the pleurae and acts as a lubricant.
Summary of Patients Results
Here is a quick summary of the patients results:
- 3 adult patients with severe pneumonia were admitted in a Wuhan hospital on December 27, 2019.
- Patient 1 was a 49 year old woman.
- Medical history was available.
- No underlying chronic medical conditions.
- Had a fever, and cough with chest discomfort on December 23, 2019.
- 4 days after onset of illness, her cough and chest discomfort worsened, but fever was reduced.
- Diagnosed with pneumonia based on a CT scan.
- She worked as a retailer in a seafood market.
- Recovered and left the hospital on January 16, 2020.
- Patient 2 was a 61 year old man.
- Medical history was available.
- Had a fever and cough on December 20, 2019.
- Had trouble breathing 7 days after the illness started.
- The illness worsened over the next 2 days, at which time a ventilator was used to artificially push air into and out of his lungs.
- Died on January 9, 2020.
- Patient 3 was a 32 year old man.
- Recovered and left the hospital on January 16, 2020.
- No biopsy or tissue samples were obtained from any of the patients.
3 patients, a small sample size, no samples taken, and still no "virus" proved anywhere.
Paper Results: Detection and Isolation of a New "Virus"
Now let's take a look at the next part of the paper results, this time on supposedly "detecting and isolating a new virus".
DETECTION AND ISOLATION OF A NOVEL CORONAVIRUS
Three bronchoalveolar-lavage samples were collected from Wuhan Jinyintan Hospital on December 30, 2019. No specific pathogens (including HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1) were detected in clinical specimens from these patients by the RespiFinderSmart22kit. RNA extracted from bronchoalveolar-lavage fluid from the patients was used as a template to clone and sequence a genome using a combination of Illumina sequencing and nanopore sequencing. More than 20,000 viral reads from individual specimens were obtained, and most contigs matched to the genome from lineage B of the genus betacoronavirus — showing more than 85% identity with a bat SARS-like CoV (bat-SL-CoVZC45, MG772933.1) genome published previously.
HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 are types of human "coronaviruses". "RNA template to clone" refers to creating DNA from a specific RNA sequence and then amplifying the DNA to make many copies. Viral reads are short DNA or RNA sequences generated after breaking down "viral" genetic material into small fragments and then using high-throughput sequencing technology to sequence the fragments.
Continuing further into the paper:
Positive results were also obtained with use of a real-time RT-PCR assay for RNA targeting to a consensus RdRp region of pan β-CoV (although the cycle threshold value was higher than 34 for detected samples). Virus isolation from the clinical specimens was performed with human airway epithelial cells and Vero E6 and Huh-7 cell lines. The isolated virus was named 2019-nCoV.
The PCR cycle threshold or Ct value is the specified maximum number of PCR cycles allowed for a positive reading to be accepted when detecting a specific DNA or RNA sequence; it can also be used to quantify the amount of DNA or RNA present in a sample. Vero E6 are a type of cell line derived from the kidney cells of an African green monkey in 1968 as the Vero 76 cell line and then cloned in 1979 to form the Vero E6 cell line; these cell lines are commonly used in "virus isolation" because they are highly permissive to "viral infection". A cell line is a population of cells that are derived from a single cell and can be maintained and grown indefinitely in culture. Huh-7 is a cell line derived from liver cancer cells of a 57-year-old Japanese male in 1982. Cloning cell lines is the process of creating genetically identical cells from a single parent cell.
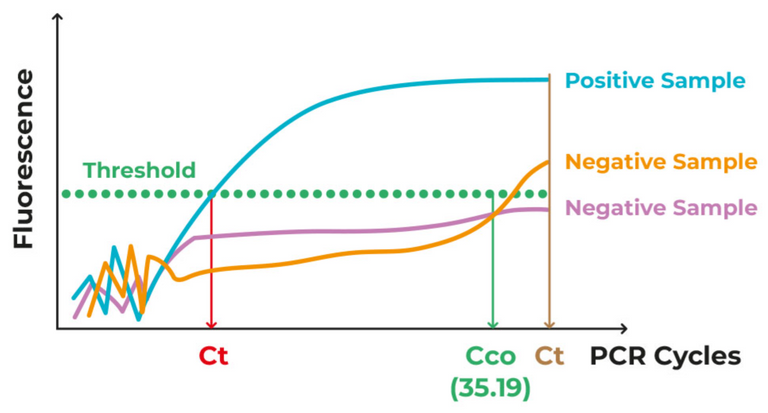
Source: Illustration of cycle threshold (Ct) for specifying a sample as positive or negative for a targeted sequence.
Continuing further in the paper:
To determine whether virus particles could be visualized in 2019-nCoV–infected human airway epithelial cells, mock-infected and 2019-nCoV–infected human airway epithelial cultures were examined with light microscopy daily and with transmission electron microscopy 6 days after inoculation. Cytopathic effects were observed 96 hours [4 days] after inoculation on surface layers of human airway epithelial cells; a lack of cilium beating was seen with light microcopy in the center of the focus (Figure 2). No specific cytopathic effects were observed in the Vero E6 and Huh-7 cell lines until 6 days after inoculation.

Figure 2. Cytopathic Effects in Human Airway Epithelial Cell Cultures after Inoculation with 2019-nCoV.
Mock-infected is a control group in which the same procedure is done but without the actual "virus". Cilium beating refers to the coordinated movement of cilia (plural of cilium).
Continuing further in the paper.
Electron micrographs of negative-stained 2019-nCoV particles were generally spherical with some pleomorphism (Figure 3). Diameter varied from about 60 to 140 nm. Virus particles had quite distinctive spikes, about 9 to 12 nm, and gave virions the appearance of a solar corona. Extracellular free virus particles and inclusion bodies filled with virus particles in membrane-bound vesicles in cytoplasm were found in the human airway epithelial ultrathin sections. This observed morphology is consistent with the Coronaviridae family.
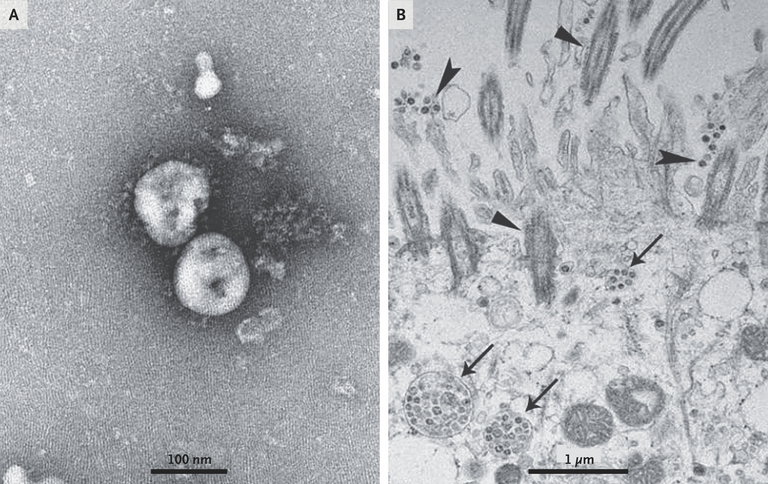
Figure 3. Visualization of 2019-nCoV with Transmission Electron Microscopy.
Negative-stained 2019-nCoV particles [from the supernatant] are shown in Panel A, and 2019-nCoV particles in the human airway epithelial cell ultrathin sections are shown in Panel B. Arrowheads indicate extracellular virus particles, arrows indicate inclusion bodies formed by virus components, and triangles indicate cilia.
Pleomorphism refers to the variability in the size and shape of cells or their nuclei. Solar or stellar corona refers to the outermost layer of a star's atmosphere, and during a total solar eclipse, it can be seen by the naked eye. A total solar eclipse occurs when the Moon completely blocks the Sun from being seen from a point on the Earth. Extracellular free virus particles are "viruses" found outside of and released by "infected cells"; these supposedly can be spread through coughing, talking, sneezing, and inhaled by others to start a "new infection". Virus inclusion bodies or viroplasm are supposedly "viral factories" where the "virus" replicates and assembles. Membrane-bound vesicles are structures within cells that are enclosed by a membrane and have various purposes, such as transporting, storing, or breaking down materials. The cytoplasm is all the material within a cell.

Total solar eclipse making the solar corona visible with the naked eye
Illustration of a solar eclipse
Summary of "New Virus" Detection and Isolation
Here is a summary of the paper results on detecting and isolating this "new virus":
- 3 airway fluid samples were collected from a Wuhan hospital on December 30, 2019.
- No specific known "viruses" were detected from these patients.
- RNA was extracted from the airway fluid samples, and assumed to be from a "virus".
- The RNA was used as a template to create DNA, which was later amplified to many copies, and sequencing technology was used to produce a genome.
- 20,000 short RNA sequences were obtained and long continuous sequences from these short and overlapping sequences (i.e. contigs) were produced and showed 85% similarity with a bat coronavirus.
- PCR tests using a cycle threshold (Ct) higher than 34 were positive when using a known sequence common among beta-coronaviruses.
- "Virus" isolation from the clinical specimens was performed on human airway epithelial cells, monkey kidney cells, and liver cancer cells from a 57-year-old Japanese man.
- The "isolated virus" was named 2019-nCoV or 2019 novel Coronavirus.
- The "virus" was then used to "infect" human airway epithelial cell cultures, with a "mock-infected" culture used as a control.
- The cultures were examined daily with optical light microscopy and then with TEM 6 days after "infection".
- Cytopathic effects were observed 96 hours (4 days) after inoculation on surface layers of human airway epithelial cells; with a lack of coordinated cilium movement was observed in some spots.
- In the monkey kidney and human liver cancer cell cultures, cytopathic effects were observed 6 days after "infection".

Figure 2: Supposed cytopathic effects (CPE) in human airway cell culture on the right image with a lack of cilium beating in the center.
- For the supernatant samples, electron microscope images of "virus" particles were presented, in which the background was stained with heavy metals.
- The particles varied in size and shape and ranged between 60 to 140 nm; supposedly with "spikes" 9 to 12 nm in size and supposedly looking like a "solar corona".
- For the embedded and heavy metal stained / fixed thin sections of human airway epithelial cell cultures, electron microscope images showed "viruses" outside of the cells, as well as within membrane-bound structures inside the cells (that supposedly act as "virus factories" in the replication and formation of "viruses").
- The observed "virus" particles supposedly belong to the Coronaviridae family.
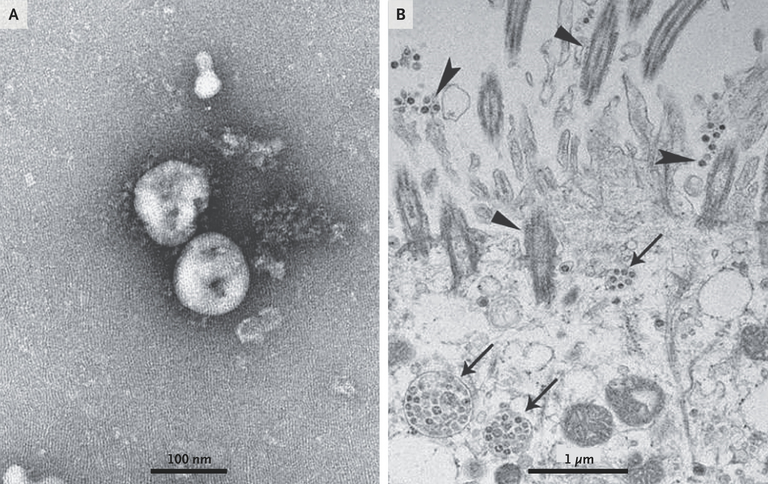
Figure 3: Negative stained supernatant on the left supposedly showing "virus" particles. On the right shows the ultrathin embedded sections: Inclusion bodies or "virus factories" within cells (arrows), "extracellular viruses" (arrowheads), and cilia (triangles).
Paper Results: Genetic Characterization of the "Virus"
This next section is still in the "detection and isolation of a novel coronavirus" results part of the paper, but it dealt specifically with genetic sequencing, so I will cover this separately.
To further characterize the virus, de novo sequences of 2019-nCoV genome from clinical specimens (bronchoalveolar-lavage fluid) and human airway epithelial cell virus isolates [presumably referring to the cell culture supernatant] were obtained by Illumina and nanopore sequencing. The novel coronavirus was identified from all three patients.
De novo sequencing refers to genetic sequencing without using any existing reference or template sequence; the process involves DNA or RNA molecules being broken into smaller fragments, sequencing those fragments, and then a computer algorithm assembles the fragments into a "complete genome sequence".
Continuing further in the paper:
Two nearly full-length coronavirus sequences were obtained from bronchoalveolar-lavage fluid (BetaCoV/Wuhan/IVDC-HB-04/2020, BetaCoV/Wuhan/IVDC-HB-05/2020|EPI_ISL_402121), and one full-length sequence was obtained from a virus isolated from a patient (BetaCoV/Wuhan/IVDC-HB-01/2020|EPI_ISL_402119). Complete genome sequences [should be 2 partial and 1 complete] of the three novel coronaviruses were submitted to GISAID (BetaCoV/Wuhan/IVDC-HB-01/2019, accession ID: EPI_ISL_402119; BetaCoV/Wuhan/IVDC-HB-04/2020, accession ID: EPI_ISL_402120; BetaCoV/Wuhan/IVDC-HB-05/2019, accession ID: EPI_ISL_402121) and have a 86.9% nucleotide sequence identity to a previously published bat SARS-like CoV (bat-SL-CoVZC45, MG772933.1) genome. The three 2019-nCoV genomes clustered together within the sarbecovirus subgenus, which shows the typical betacoronavirus organization: a 5′ untranslated region (UTR), replicase complex (orf1ab), S gene, E gene, M gene, N gene, 3′ UTR, and several unidentified nonstructural open reading frames.
GISAID or Global Initiative on Sharing All Influenza Data is a world wide database for genomic data on influenza "viruses". Influenza or "the flu" is supposedly an infectious disease caused by "influenza viruses". The genetic organization mentioned above are the supposed regions of the genome that are either noncoding or code for specific functions of the "virus", such as replication or producing the "spike" protein.
Continuing further in the paper:
Although 2019-nCoV is similar to some betacoronaviruses detected in bats (Figure 4), it is distinct from SARS-CoV and MERS-CoV. The three 2019-nCoV coronaviruses from Wuhan, together with two bat-derived SARS-like strains, ZC45 and ZXC21, form a distinct clade. SARS-CoV strains from humans and genetically similar SARS-like coronaviruses from bats collected from southwestern China formed another clade within the subgenus sarbecovirus. Since the sequence identity in conserved replicase domains (ORF 1ab) is less than 90% between 2019-nCoV and other members of betacoronavirus, the 2019-nCoV — the likely causative agent of the viral pneumonia in Wuhan — is a novel betacoronavirus belonging to the sarbecovirus subgenus of Coronaviridae family.
Replicase domains (ORF 1ab) are a set of conserved regions in the genome of coronaviruses that encode for proteins involved in "viral" replication.
Continuing further in the paper:
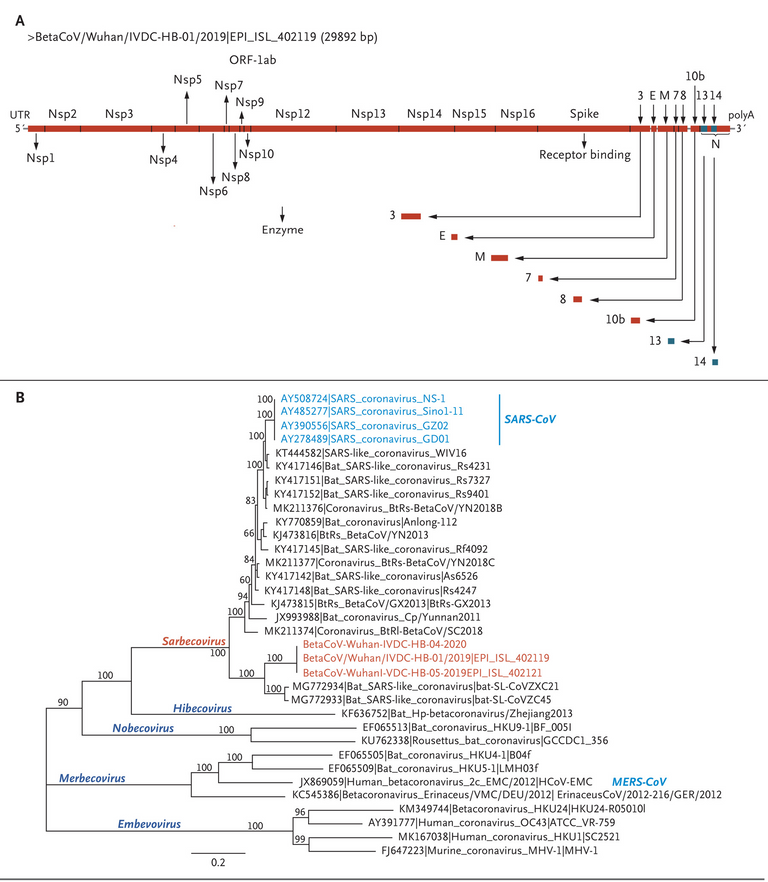
Figure 4. Schematic of 2019-nCoV and Phylogenetic Analysis of 2019-nCoV and Other Betacoronavirus Genomes.
Shown are a schematic of 2019-nCoV (Panel A) and full-length phylogenetic analysis of 2019-nCoV and other betacoronavirus genomes in the Orthocoronavirinae subfamily (Panel B).
Note the clade with the BetaCoVs (the supposed "novel COVID virus") and Bat_SARS-like_coronaviruses.
Summary of the Genetic Characterization of the "Virus"
Let's go over a summary of the paper results section on genetic characterization of the "virus":
- Genetic sequencing of the airway fluid samples and the cell culture supernatant obtained the "virus" genome.
- The "virus" genome was identified from all 3 patients.
- 2 nearly full-length "virus" sequences were obtain from the lung fluid samples.
- 1 full-length "virus" sequence was obtained from the cell culture supernatant.
- The 3 "virus" genome sequences were sent to the GISAID global flu "virus" database.
- The genome sequences had a 86.9% nucleotide sequence identity to a previously published "bat coronavirus" genome.
- The 3 "virus" genomes form a distinct clade or grouping with less than 90% similarity of genetic sequences (genes) that code for proteins involved in "viral" replication.
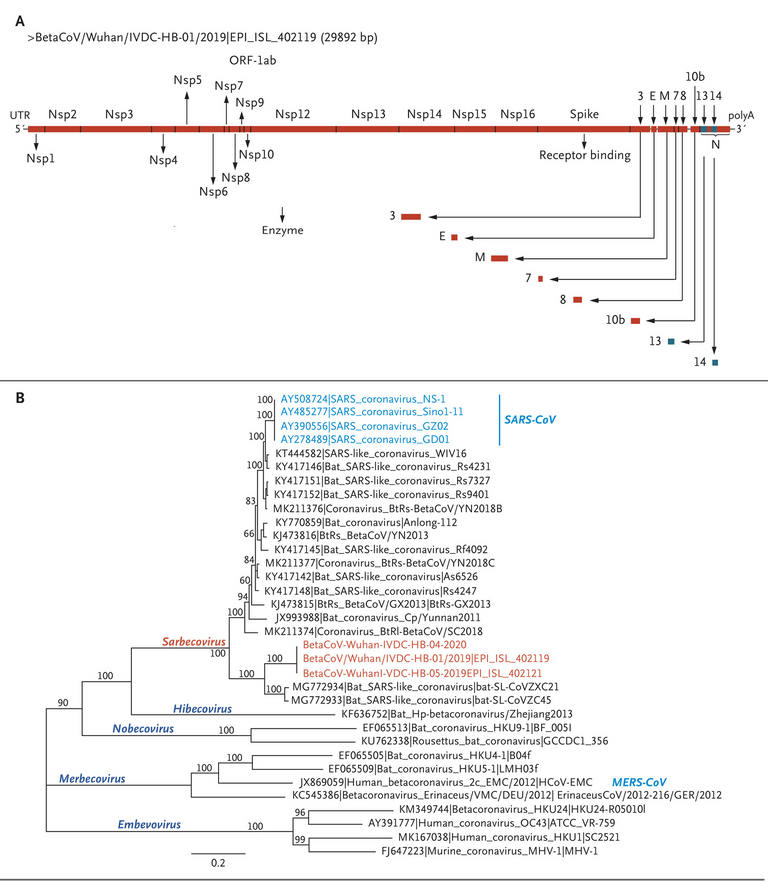
Figure 4: The red BetaCoVs are the grouping for the supposed "novel COVID virus".
Paper Discussion
Now let's go over the paper's discussion section to see how they summarize their "findings".
We report a novel CoV (2019-nCoV) that was identified in hospitalized patients in Wuhan, China, in December 2019 and January 2020. Evidence for the presence of this virus includes identification in bronchoalveolar-lavage fluid in three patients by whole-genome sequencing, direct PCR, and culture. The illness likely to have been caused by this CoV was named “novel coronavirus-infected pneumonia” (NCIP). Complete genomes were submitted to GISAID. Phylogenetic analysis revealed that 2019-nCoV falls into the genus betacoronavirus, which includes coronaviruses (SARS-CoV, bat SARS-like CoV, and others) discovered in humans, bats, and other wild animals.15 We report isolation of the virus and the initial description of its specific cytopathic effects and morphology.
Direct PCR refers to using PCR directly on the sample without prior DNA extraction or purification steps.
Continuing further into the paper:
Molecular techniques have been used successfully to identify infectious agents for many years. Unbiased, high-throughput sequencing is a powerful tool for the discovery of pathogens.14,16 Next-generation sequencing and bioinformatics are changing the way we can respond to infectious disease outbreaks, improving our understanding of disease occurrence and transmission, accelerating the identification of pathogens, and promoting data sharing. We describe in this report the use of molecular techniques and unbiased DNA sequencing to discover a novel betacoronavirus that is likely to have been the cause of severe pneumonia in three patients in Wuhan, China.
Bioinformatics is the field that develops methods and software tools for understanding big sets of biological data.
Continuing further in the paper:
Although establishing human airway epithelial cell cultures is labor intensive, they appear to be a valuable research tool for analysis of human respiratory pathogens.13 Our study showed that initial propagation of human respiratory secretions onto human airway epithelial cell cultures, followed by transmission electron microscopy and whole genome sequencing of culture supernatant, was successfully used for visualization and detection of new human coronavirus that can possibly elude identification by traditional approaches.
Further development of accurate and rapid methods to identify unknown respiratory pathogens is still needed. On the basis of analysis of three complete genomes obtained in this study, we designed several specific and sensitive assays targeting ORF1ab, N, and E regions of the 2019-nCoV genome to detect viral RNA in clinical specimens. The primer sets and standard operating procedures have been shared with the World Health Organization and are intended for surveillance and detection of 2019-nCoV infection globally and in China. More recent data show 2019-nCoV detection in 830 persons in China.17
Although our study does not fulfill Koch’s postulates, our analyses provide evidence implicating 2019-nCoV in the Wuhan outbreak.
Koch's postulates are 4 criteria designed to establish a causal link between a microbe and a disease, and are listed below.
- The microorganism must be found in abundance in all organisms suffering from the disease but should not be found in healthy organisms.
- The microorganism must be isolated from a diseased organism and grown in pure culture [containing only a population of that microbe].
- The cultured microorganism should cause disease when introduced into a healthy organism.
- The microorganism must be re-isolated from the inoculated, diseased experimental host and identified as being identical to the original specific causative agent.
Note that the postulates are not strictly followed anymore, and terms such as "asymptomatic carriers" (carrying the microbe but not the disease) and "viruses" which require host cells to "grow" are assumed in mainstream science.
Continuing further in the paper:
Additional evidence [presumably yet to be done] to confirm the etiologic [cause of disease] significance of 2019-nCoV in the Wuhan outbreak include identification of a 2019-nCoV antigen in the lung tissue of patients by immunohistochemical analysis, detection of IgM and IgG antiviral antibodies in the serum samples from a patient at two time points to demonstrate seroconversion, and animal (monkey) experiments to provide evidence of pathogenicity. Of critical importance are epidemiologic investigations to characterize transmission modes, reproduction interval, and clinical spectrum resulting from infection to inform and refine strategies that can prevent, control, and stop the spread of 2019-nCoV.
2019-nCoV antigen refers to the proteins or regions of proteins (such as the "spike protein") on the "virus" that can serve as antigens and "stimulate the immune system". Immunohistochemical (IHC) analysis is a technique used to visualize and detect specific proteins or antigens within tissue samples. Immunoglobulin M (IgM) is the largest antibody, and first antibody to appear in the response to initial exposure to an antigen. Immunoglobulin G (IgG) is the most common type of antibody found in blood circulation and represents approximately 75% of serum antibodies in humans. Seroconversion is the development of specific antibodies in the blood serum as a result of "infection" or "immunization" (including vaccination). Immunization is the assumed process of strengthening an individual's "immune system" after an initial exposure to an "infectious agent". Pathogenicity is a measure of how effective a pathogen is at "causing disease". Transmission mode is the specific way a disease or pathogen is "transmitted" to another person or organism. Reproduction level is the average time it takes for an infected individual to "transmit the disease to others". Clinical spectrum is the full range of signs, symptoms, and outcomes that can occur as a result of a "disease" or condition.
Summary of Paper Discussion
Let's go over a summary of the paper's discussion section:
- A "novel CoV" was identified in hospital patients in December 2019 and January 2020.
- Evidence for this "new virus" includes "identification" in airway fluid samples by the following methods:
- Whole-genome sequencing.
- PCR directly on the samples.
- Cell cultures from the samples.
- "Complete" genomes were submitted to the GISAID and the "new virus" falls into the "betacoronavirus" classification.
- Modern genetic sequencing and big data analysis technology is "accelerating the identification of pathogens" such as this "new virus" that is "likely" to have been the cause of severe pneumonia in 3 patients in Wuhan, China.
- Human lung fluid samples, with the assumption that they "contain a new virus", were used to "infect" human airway epithelial cell cultures.
- "Virus" particles were assumed to be visualized via an electron microscope.
- Whole genome sequencing was performed on the cell culture supernatant.
- From the 3 "complete" genomes, the authors developed techniques to identify this "new virus" by targeting specific sequences from these "genomes" in the targeting of "viral RNA" in future patients.
- The sequences and procedures were shared with the WHO for the surveillance and detection of this "new virus" globally.
- As of the paper's publishing, 830 people in China "tested positive" for these targeted sequences.
- The authors admit their paper does not fulfill Koch's postulates of a more concrete link of microbe-to-disease causation.
- The authors instead claim their study "implicates" the "new virus" in the Wuhan "outbreak".
- Additional experiments need to be done to confirm the disease-causing claim of this "new virus", and these are:
- Identify proteins on the "virus" that serve as antigens in the lung tissue of patients.
- Detect corresponding antibodies or proteins in the blood of patients that form to "identify and neutralize" the "virus" antigens.
- Animal (monkey) experiments to demonstrate the "virus" actually causes disease.
- The paper concludes by highlighting the importance of "epidemiological investigations" to characterize the transmission and clinical symptoms of this "disease" in order to refine strategies to "stop the spread of this new virus".
MES Summary
This video has served as a study of mainstream virology in general for me, and I hope it has for you as well.
While the video covers the paper in its entirety, much of it discusses assumptions, theories, and narratives beyond the specific experiments actually performed in the paper.
So now, to better grasp the key claims of the paper, I will summarize only the main parts of the paper's most important claim, which is the assumption of a "new disease-causing virus":
- ChatGPT says the following paper is the main paper on the isolation of the "virus" that causes COVID-19.
- Paper Link: https://www.nejm.org/doi/full/10.1056/NEJMoa2001017
- A Novel Coronavirus from Patients with Pneumonia in China, 2019
- By Zhu et al (Latin for "and others")
- Published on January 24, 2020.
- Updated on January 29, 2020.
- Appears in the February 20, 2020 issue of The New England Journal of Medicine (NEJM).
- On December 31, 2019 the China CDC sent a rapid response team to Wuhan to investigate reported clusters of patients with pneumonia (or inflammation of the lungs) of "unknown cause", and had recently been to or in contact with someone that had been to the Huanan Seafood Market in Wuhan.
- This paper reports the results of the investigation.
- The chest radiographs of a patient, whom no samples were taken from, with pneumonia and assumed to be from the "new virus" is shown below:
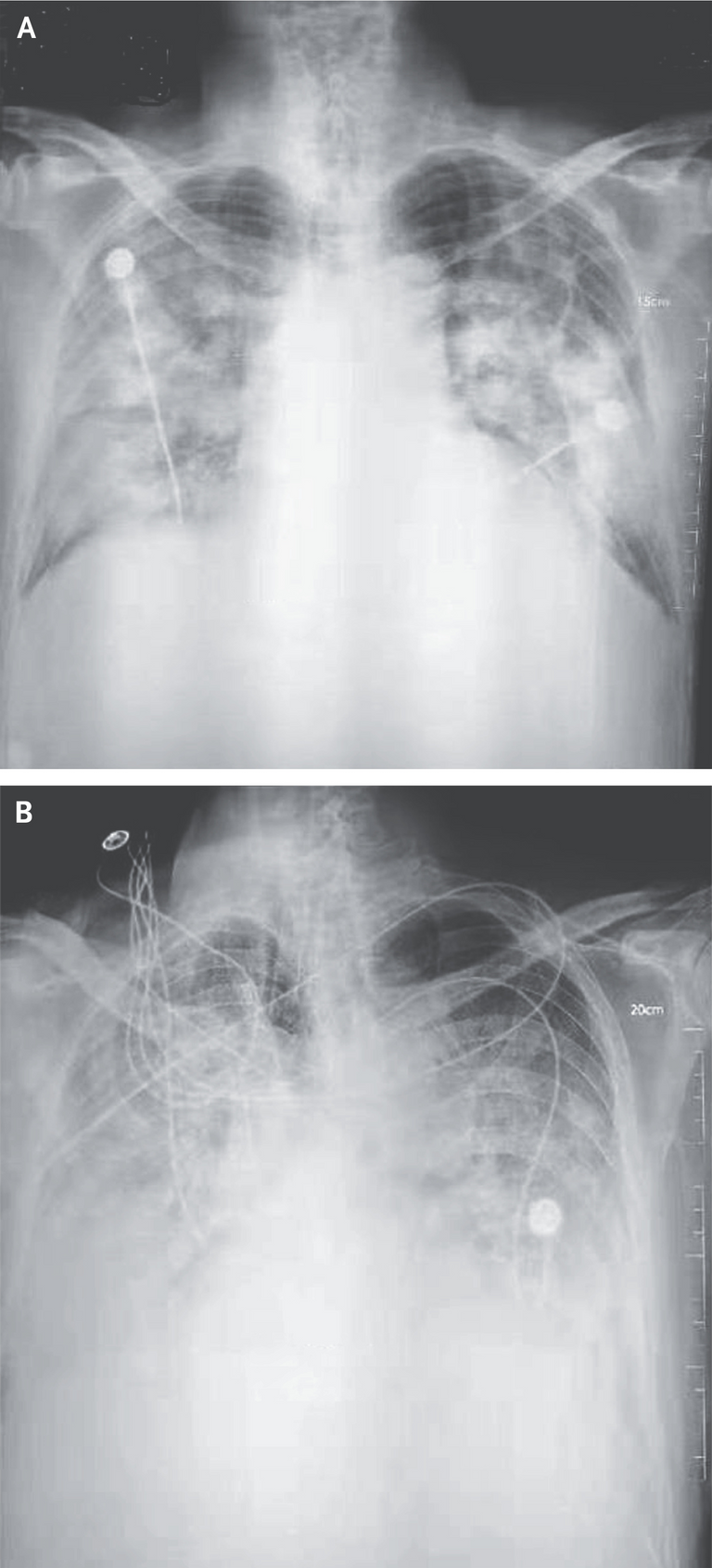
Figure 1: Patient 2 on days 8 and 11 after start of the illness. Mechanical ventilation or assisted breathing was performed between the time period of the two images. The cloudy or hazy areas (more present in the bottom image) supposedly indicate accumulation of fluid around the lungs.
- 4 lower respiratory tract (throat / lungs region) samples were collected from the unknown-cause pneumonia patients who were in Wuhan on December 21, 2019 or later and had been present in the market.
- 7 samples were collected from known-cause pneumonia patients from Beijing to be used as control samples.
- Nucleic acids were extracted from both sets of samples.
- PCR (or DNA replication) tests were done on the nucleic acids to test for known sequences from 22 "viruses" and 4 bacteria; and presumably they weren't found in the Wuhan samples.
- Modern whole genome sequencing technology was used to "discover" sequences not identifiable by the above PCR methods.
- RT-PCR (RNA conversion to DNA then DNA replication) assay was used to detect "viral RNA" by targeting a specific genetic sequence that is common to many "betacoronaviruses".
- After discovering "unknown sequences" and "viral RNA", the next step was to "isolate the virus".
- Respiratory samples were collected from the unknown-cause pneumonia patients.
- Chemical preservatives were added (possibly including fetal bovine serum (FBS)) to the samples.
- Samples were centrifuged to remove "cellular debris".
- The supernatant was collected.
- Passage 1: The supernatant was used to "infect" human airway epithelial cells obtained from lung cancer surgery patients.
- The airway cells were initially grown on a plastic surface in an air-liquid interface (bottom in contact with liquid and top with air) for 4 to 6 weeks.
- Prior to "supernatant infection", the top parts of the airway cell cultures were washed 3 times with a water-based salt solution (phosphate-buffered saline (PBS)).
- 0.15 mL of supernatant was placed (aka "infected") on the top surface of the cell culture.
- The "supernatant-infected cell culture" was then placed in an incubator at 37°C with 5% CO2 in an air-liquid interface.
- After 2 hours of incubation, "unbound virus" was removed by washing with 0.50 mL of PBS for 10 minutes.
- Every 48 hours, 0.15 mL of PBS was added to the top surface (apical) of the airway cells.
- After 10 minutes of incubation at 37°C the apical samples were harvested (removed).
- The removed apical (top surface) samples were diluted at a ratio of 1 to 3 to a new cell culture and monitored daily for:
- Cytopathic effects (cell breakdown / cell death) via an optical light microscope.
- "Viral nucleic acid" in the supernatant via RT-PCR.

Figure 2: "Mock-infected" (which the paper doesn't elaborate any further) on the left and "new virus infected" human airway epithelial (HAE) cell cultures on the right. Visualized with an optical light microscope. Cytopathic effects (CPE) were supposedly observed 4 days after "infection" and the arrow points to a lack of coordinated movement from cilia (hair-like structures on the surface of epithelial cells that move mucus and trapped particles out of the respiratory tract).
- The supernatant in the last step was then removed and used to "infect" another airway cell culture; then repeated again for a total of 3 Passages.
- After these 3 passages, supernatant from the airway cell cultures that showed "cytopathic effects" were then collected.
- The supernatant was then "virus inactivated" for at least 2 hours by the chemical preservative paraformaldehyde, which "fixes" or locks molecules in place preventing further changes (aka prevents "virus replication").
- Then, the supernatant was ultracentrifuged to sediment "virus" particles (this sediment is called "enriched supernatant").
- The enriched supernatant was then placed on thin grids stained with heavy metals (to provide contrast for electron microscope imaging).
- Similarly, after 3 passages, the airway cell cultures themselves (not the supernatant) that showed "cytopathic effects" were collected.
- The airway cell cultures were fixed with the chemical preservatives paraformaldehyde and glutaraldehyde.
- Then they were fixed with the heavy metal preservative osmium tetroxide.
- Then they were dehydrated with ethanol alcohol, which penetrates cell membranes and replaces water molecules within the cells.
- Then they were embedded in the resin PON812.
- Ultrathin 80 nm sections were cut from the resin block and stained with the heavy metals uranyl acetate and lead citrate.
- The negative stained grids (containing the supernatant) and ultrathin sections (containing the airway cell cultures) were observed with an electron microscope.

Figure 3: Electron microscope images. A: "Virus particles" from the supernatant. B: Airway cell cultures: Arrowheads indicate "extracellular viruses", arrows indicate inclusion bodies or "virus factories" within cells, and triangles indicate cilia.
The "virus" particles (termed "virions") supposedly show "spikes" on the outer surface, and supposedly give the appearance of a solar corona… #Supposedly
- RNA was extracted from the patient airway fluid samples and cell culture supernatants.
- This RNA was assumed to be from a "virus".
- This RNA was used as a template for modern sequencing technology to produce a "virus genome".
- This "novel virus genome" was identified in "all 3 patients".
- The 3 genome sequences were sent to the GISAID (Global Initiative on Sharing All Influenza Data) global "flu" virus database.
- Statistical and computational analysis were performed on the likely "evolutionary history" of the "virus" genome.
- The 3 genomes have a 86.9% similarity with a previously published "bat coronavirus" genome.

Figure 4: The red BetaCoVs are the grouping for the supposed "novel COVID virus".
MES Conclusions
After sifting through this paper with a fine-toothed comb, the only thing clear is that there is nothing clear about the paper, the methods, or virology in general.
A handful of patients were assumed to have a "viral" infection.
A handful of respiratory samples were obtained and assumed to contain a "virus".
Genetic sequencing aka "listing of molecules" and comparing with public databases of other genetic sequences was used as the basis for identifying this "new virus".
Respiratory samples were heavily processed, centrifuged, mixed with chemical preservatives, grown and regrown in cell cultures derived from cancer patients, stained with heavy metals, washed with saline solutions, embedded in resin, cut in ultrathin sections, and placed in high-powered electron microscopes.
Optical light microscopy images showed non-persuasive information, to say the least.
Electron microscope images show dead particles which are assumed to be "virions".
Besides requiring further research into the "genetic sequencing" science being performed, there is not much of any significance presented in this paper; and much less what would be expected to declare a "pandemic" and lockdown, mask up, and vax up the world.
Interestingly, there some very peculiar timing of this whole COVID psychological operation (PsyOp), and PsyOp it most certainly is:
- December 1, 2019: China's new strict vaccine law goes into effect.
- December 1, 2019: "Patient Zero" started to have symptoms.
- December 31, 2019: China CDC sends a rapid response team to Wuhan.
And of course, the paper ends with the now common place brainwashing propaganda urging the public and governments to "prevent, control, and stop the spread of 2019-nCoV"…
Stay tuned for #MESScience 5…

https://reddit.com/r/AMAZINGMathStuff/comments/141k1qz/extensive_review_of_the_main_paper_on_covid19/
The rewards earned on this comment will go directly to the people sharing the post on Reddit as long as they are registered with @poshtoken. Sign up at https://hiveposh.com.
This already paid-out post was just upvoted by @mikewick77.
This post was created to allow this voter the oportunity to still upvote this content IF (s)he deems this content to be high quality.
Please don't use this post to self-upvote. You will get blacklisted if you do. And don't use it to upvote low quality content
This is a hive-archeology proxy comment meant as a proxy for upvoting good content that is past it's initial pay-out window.
Pay-out for this comment is configured as followed: