Genetic Sequencing technologies is still evolving?
A while ago I wrote an article about how any person can have their Genome sequenced or use other genotyping assays with private companies that charge accessible prices so anyone can check if there is any known variants for certain diseases in here . I still didn't have my genome sequenced, my daughter, my wife and I tried 23andMe only which uses the SNP array technology, which brought us interesting information about our genomes, and our inheritance and maybe opened a little bit of our eyes for some health possibilities in the future.
Since I started to work in the bioinformatics area, more precisely dealing with sequencing data to analyze and extract important information, it changed a bit. I started to work in the are in 2009, 8 years after the first draft of the human genome was published in Nature and Science journals in 2001. At that time, some sequencing platforms were popular, and today they aren't as much. People were using the platform ABI Solid produced by Life Technologies to generate small fragments ( or reads how they are called in the area) of 50 basepairs, and taking in account that the human genome sequence has around 3 billion base pairs, imagine how many fragments we would need to cover all this genome. Another interesting thing is that it used a method called color space to identify the nucleotides which nowadays were abandoned, probably because it was a little bit complex.
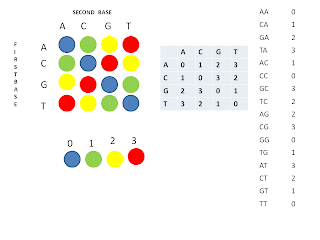
Colour space method [1]
Another platform that was pretty used mostly with people studying bacteria was the Roche 454 sequencing platform, this method generating at that time what was considered the longest read could be up to 600 bp, and for people dealing with small genomes, like in bacterias, was interesting since it could cover more of the genome with less reads. However, the throughput was lower compared to SOLID, that's why if you did a sequencing experiment with 454 in human samples, you would have a small number of reads , so less of the genome covered. The 454 technology also brought one innovation, the pyrosequencing, which is nothing more that using a common nature reaction to identify when a new ligation between nucleotides happens: the one that happens inside fireflies the release of luciferin that produces light! So every time that an A or T or C or G ligated to read a sequence that was beginning to be formed, luciferin is released and sensors detect light.
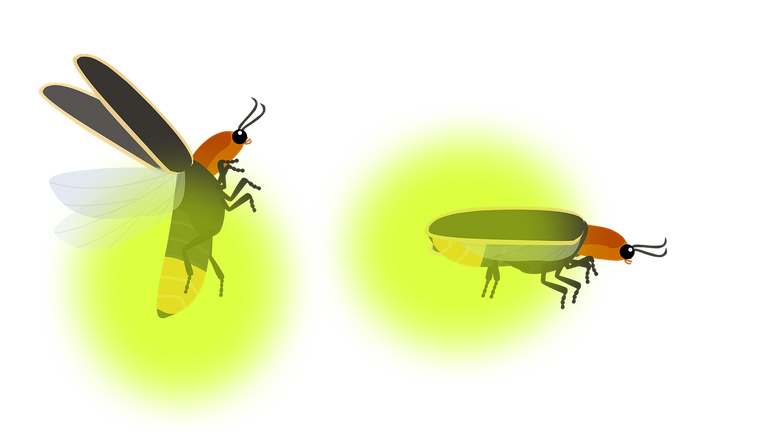
Firefly used as reference in the pyrosequencing of 454 [2]
At that time another company started to grow a little bit in the field called Illumina. This company bought an old sequencing company called Solexa and started to evolve an old method, the Sanger sequencing created in 1977. Illumina implemented the same method of identification of nucleotides based on their fluorescence , for each nucleotide you find a fluorescent marker attached, and when the nucleotide ligate to the read sequence being formed, it releases this fluorescent molecule that is detected by a sensor. This technology at that time generated read lengths of around 100 base pairs. But with throughput of 600 Gb of data, the double of ABI solid with their 300 Gb of data more than 600 more than 454 with their 0.7 Gb of data generated. These 3 platforms continued to grow, but for sure Illumina grew much more. Nowadays the company has 5 different platforms with different throughputs and costs, so depending of your usage they have the correct cost/benefit for your data generation and the read length raised a bit to 150bp, and the top machine called NovaSeq produces 6,000 Gb of data.
One year after my beginning a company called Ion Torrent, a subsidiary from Life Technologies, developed a different sequencing method that promised a cheap and efficient way to sequence, using the power of natural ligation. Instead of using a sophisticated method of ligation detection of nucleotides, the method would detect the natural release of Hydrogen protons released naturally with a pH sensor. Thermo Fisher, an important biotechnology company bought Life Technologies and this technology together, however, nowadays this technology showed some deficiencies, such as a not very precise nucleotide detection. Unfortunately, the Ion Torrent technology has being dropped its usage in big laboratories in the world.
Illumina grew, however the 454 technology left opened the opportunities of new long read platforms and technologies to be developed. Two examples appeared less than 10 years ago: Pacific Biosciences and Nanopore Minion. Both technologies brought the concept of "natural sequencing". All the previous technologies break the genetic material and sequence by fragmented sequences of genetic material. In these two technologies, the entire DNA or RNA is being read simultaneously producing long reads which can reach thousands of nucleotides. Nanopore Minion for example was born in Oxford in a university initiative, and it uses a pore ina membrane where the genetic material passes by and sensors detect by electric conductivity which nucleotide just passed by. Researchers studying small genomes organisms are the more beneficiary from these technologies since, the genome is less complex and the technology still generates some errors during sequencing. However, during the COVID pandemic it was broadly used to identify new variants.
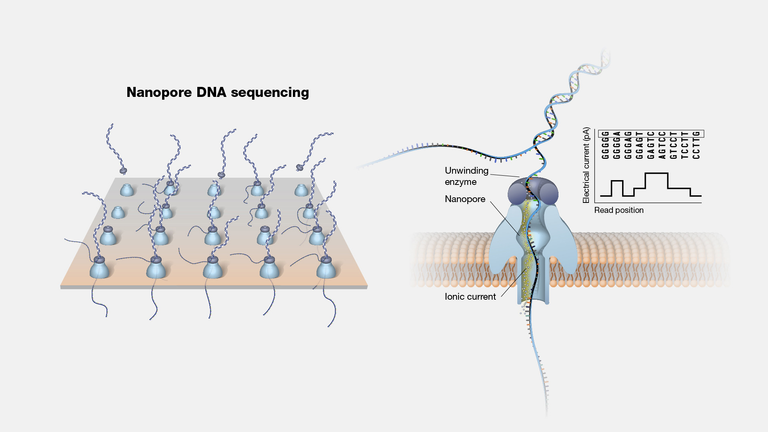
Nanopore sequencing, the genetic material passing by the nanopores of the technology [3]
In the last 14 years that I joined this world, technologies started to be less used, new technologies appeared, and more detailed data started to be generated. Cost is still challenging, but it looks like it is going down every year with the new technologies. Maybe in a couple of years, we will have our finger stung and in less than one hour have our genome sequenced.

banner created in canvas
https://twitter.com/1451373011018338304/status/1638665992577380354
The rewards earned on this comment will go directly to the people( @gwajnberg ) sharing the post on Twitter as long as they are registered with @poshtoken. Sign up at https://hiveposh.com.
Thanks for your contribution to the STEMsocial community. Feel free to join us on discord to get to know the rest of us!
Please consider delegating to the @stemsocial account (85% of the curation rewards are returned).
You may also include @stemsocial as a beneficiary of the rewards of this post to get a stronger support.
Congratulations @gwajnberg! You have completed the following achievement on the Hive blockchain And have been rewarded with New badge(s)
Your next target is to reach 17000 upvotes.
You can view your badges on your board and compare yourself to others in the Ranking
If you no longer want to receive notifications, reply to this comment with the word
STOPCheck out our last posts: